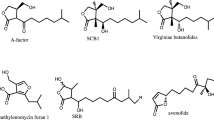
Abstract
The bacterial genus Streptomyces is well characterized by an ability to produce a wide variety of secondary metabolites including antibiotics, anticancer agents, antiviral drugs, herbicides, pesticides, insecticides, and immunosuppressants. Their biosynthesis is strictly controlled by small diffusible signaling molecules that constitute a signaling molecule/receptor regulatory systems. These signaling molecule/receptor regulatory systems are extensively studied in various Streptomyces species to understand the regulatory function for secondary metabolite production. This chapter focuses on the Streptomyces signaling molecules in view of structural classification, distribution, and biosynthesis. In addition, this chapter reviews the signaling molecule-dependent regulatory systems including the receptor, the target activator, and the pseudo-receptor that are responsible for controlling secondary metabolite production in Streptomyces. Application for activation of silent secondary metabolites by manipulation of regulatory systems is also briefly described.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ahmed Y, Rebets Y, Tokovenko B, Brotz E, Luzhetskyy A (2017) Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci Rep 7:9784
Aigle B, Pang X, Decaris B, Leblond P (2005) Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J Bacteriol 187:2491–2500
Ajithkumar V, Karuppasamy K, Prasad R (2013) Regulation of daunorubicin biosynthesis in Streptomyces peucetius—feed forward and feedback transcriptional control. J Basic Microbiol 2013(53):636–644
Arakawa K (2014) Genetic and biochemical analysis of the antibiotic biosynthetic gene clusters on the Streptomyces linear plasmid. Biosci Biotechnol Biochem 78:183–189
Arakawa K (2018) Manipulation of metabolic pathways controlled by signaling molecules, inducers of antibiotic production, for genome mining in Streptomyces spp. Antonie Van Leeuwenhoek 111:743–751
Arakawa K, Mochizuki S, Yamada K, Noma T, Kinashi H (2007) γ-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153:1817–1827
Arakawa K, Tsuda N, Taniguchi A, Kinashi H (2012) The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. ChemBioChem 13:1447–1457
Arias P, Fernandez-Moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968
Aroonsri A, Kitani S, Hashimoto J, Kosone I, Izumikawa M, Komatsu M, Fujita N, Takahashi Y, Shin-ya K, Ikeda H, Nihira T (2012) Pleiotropic control of secondary metabolism and morphological development by KsbC, a butyrolactone autoregulator receptor homologue in Kitasatospora setae. Appl Environ Microbiol 78:8015–8024
Bate N, Butler AR, Gandecha AR, Cundliffe E (1999) Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem Biol 6:617–624
Bate N, Stratigopoulos G, Cundliffe E (2002) Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol Microbiol 43:449–458
Biarnes-Carrera M, Breitling R, Takano E (2015) Butyrolactone signalling circuits for synthetic biology. Curr Opin Chem Biol 28:91–98
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Bunet R, Mendes MV, Rouhier N, Pang X, Hotel L, Leblond P, Aigle B (2008) Regulation of the synthesis of the angucyclinone antibiotic alpomycin in Streptomyces ambofaciens by the autoregulator receptor AlpZ and its specific ligand. J Bacteriol 190:3293–3305
Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B (2011) Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J Bacteriol 193:1142–1153
Chen Y, Wendt-Pienkowski E, Shen B (2008) Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J Bacteriol 190:5587–5596
Chen YW, Liu XC, Lv FX, Li P (2019) Characterization of three regulatory genes involved in enduracidin biosynthesis and improvement of enduracidin production in Streptomyces fungicidicus. J Appl Microbiol 127:1698–1705
Corre C, Song L, O’Rourke S, Chater KF, Challis GL (2008) 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc Natl Acad Sci U S A 105:17510–17515
Cundliffe E (2008) Control of tylosin biosynthesis in Streptomyces fradiae. J Microbiol Biotechnol 18:1485–1491
Du D, Katsuyama Y, Onaka H, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y (2016) Production of a novel amide-containing polyene by activating a cryptic biosynthetic gene cluster in Streptomyces sp. MSC090213JE08. Chembiochem 17:1464–1471
Fernandez-Moreno MA, Caballero JL, Hopwood DA, Malpartida F (1991) The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769–780
Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E (2010) Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156:2343–2353
Handel F, Kulik A, Mast Y (2020) Investigation of the autoregulator-receptor system in the pristinamycin producer Streptomyces pristinaespiralis. Front Microbiol 11:580990
Hara O, Beppu T (1982) Mutants blocked in streptomycin production in Streptomyces griseus—the role of A-factor. J Antibiot 35:349–358
Higashi T, Iwasaki Y, Ohnishi Y, Horinouchi S (2007) A-Factor and phosphate depletion signals are transmitted to the grixazone biosynthesis genes via the pathway-specific transcriptional activator GriR. J Bacteriol 189:3515–3524
Horinouchi S, Beppu T (2007) Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Prod Jpn Acad Ser B Phys Biol Sci 83:277–295
Hsiao NH, Nakayama S, Merlo ME, de Vries M, Bunet R, Kitani S, Nihira T, Takano E (2009) Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem Biol 16:951–960
Kawauchi R, Akashi T, Kamitani Y, Sy A, Wangchaisoonthorn U, Nihira T, Yamada Y (2000) Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol Microbiol 36:302–313
Khokhlov AS, Tovarova II, Borisova LN, Pliner SA, Shevchenko LN, Kornitskaia EI, Ivkina NS, Rapoport IA (1967) The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini. Dokl Akad Nauk SSSR 177:232–235
Kinoshita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y (1997) Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J Bacteriol 179:6986–6993
Kitani S, Yamada Y, Nihira T (2001) Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a Novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J Bacteriol 183:4357–4363
Kitani S, Miyamoto KT, Takamatsu S, Herawati E, Iguchi H, Nishitomi K, Uchida M, Nagamitsu T, Omura S, Ikeda H, Nihira T (2011) Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc Natl Acad Sci U S A 108:16410–16415
Kondo K, Higuchi Y, Sakuda S, Nihira T, Yamada Y (1989) New virginiae butanolides from Streptomyces virginiae. J Antibiot 42:1873–1876
Kong D, Wang X, Nie J, Niu G (2019) Regulation of antibiotic production by signaling molecules in Streptomyces. Front Microbiol 10:2927
König CC, Scherlach K, Schroeckh V, Horn F, Nietzsche S, Brakhage AA, Hertweck C (2013) Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem 14:938–942
Koomsiri W, Inahashi Y, Leetanasaksakul K, Shiomi K, Takahashi YK, O Mura S, Samborskyy M, Leadlay PF, Wattana-Amorn P, Thamchaipenet A, Nakashima T (2019) Sarpeptins A and B, lipopeptides produced by Streptomyces sp. KO-7888 overexpressing a specific SARP regulator. J Nat Prod 82:2144–2151
Krause J, Handayani I, Blin K, Kulik A, Mast Y (2020) Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in Streptomycetes. Front Microbiol 11:225
Kunitake H, Hiramatsu T, Kinashi H, Arakawa K (2015) Isolation and biosynthesis of an azoxyalkene compound produced by a multiple gene disruptant of Streptomyces rochei. ChemBioChem 16:2237–2243
Li X, Wang J, Li S, Ji J, Wang W, Yang K (2015) ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci Rep 5:14831
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Mol Biol Rev 77:112–143
Madduri K, Hutchinson CR (1995) Functional characterization and transcriptional analysis of a gene cluster governing early and late steps in daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol 177:3879–3884
Martin J, Liras P (2020) The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in Streptomyces and related actinobacteria. Front Microbiol 10:3120
Mast Y, Guezguez J, Handel F, Schinko E (2015) A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl Environ Microbiol 81:6621–6636
Matselyukh B, Mohammadipanah F, Laatsch H, Rohr J, Efremenkova O, Khilya V (2015) N-methylphenylalanyl-dehydrobutyrine diketopiperazine, anA-factor mimic that restores antibiotic biosynthesis and morphogenesis in Streptomyces globisporus 1912-B2 and Streptomyces griseus 1439. J Antibiot 68:9–14
Matsuno K, Yamada Y, Lee CK, Nihira T (2004) Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch Microbiol 181:52–59
Mingyar E, Feckova L, Novakova R, Bekeova C, Kormanec J (2015) A gamma-butyrolactone autoregulator-receptor system involved in the regulation of auricin production in Streptomyces aureofaciens CCM 3239. Appl Microbiol Biotechnol 99:309–325
Misaki Y, Yamamoto S, Suzuki T, Iwakuni M, Sasaki H, Takahashi Y, Inada K, Kinashi H, Arakawa K (2020) SrrB, a pseudo-receptor protein, acts as a negative regulator for lankacidin and lankamycin production in Streptomyces rochei. Front Microbiol 11:1089
Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Schroeckh V, Brakhage AA (2015) Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 6:299
Nguyen TB, Kitani S, Shimma S, Nihira T (2018) Butenolides from Streptomyces albus J1074 act as external signals to stimulate avermectin production in Streptomyces avermitilis. Appl Environ Microbiol 84:e2791–e2717
Niu G, Chater KF, Tian Y, Zhang J, Tan H (2016) Specialized metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol Rev 40:554–573
Ohnishi Y, Kameyama S, Onaka H, Horinouchi S (1999) The A-factor regulatory cascade leading to streptomycin production in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol 34:102–111
Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439
Okamoto S, Nakamura K, Nihira T, Yamada Y (1995) Virginiae butanolide binding protein from Streptomyces virginiae. Evidence that VbrA is not the virginiae butanolide binding protein and reidentification of the true binding protein. J Biol Chem 270:12319–12326
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S (1995) Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol 177:6083–6092
Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF (1997) A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J Bacteriol 179:2053–2059
Recio E, Colinas A, Rumbero A, Aparicio JF, Martín JF (2004) PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J Biol Chem 279:41586–41593
Rehakova A, Novakova R, Feckova L, Mingyar E, Kormanec J (2013) A gene determining a new member of the SARP family contributes to transcription of genes for the synthesis of the angucycline polyketide auricin in Streptomyces aureofaciens CCM 3239. FEMS Microbiol Lett 346:45–55
Rutledge PJ, Challis GL (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13:509–523
Salehi-Najafabadi Z, Barreiro C, Rodríguez-García A, Cruz A, López GE, Martín JF (2014) The gamma-butyrolactone receptors BulR1 and BulR2 of Streptomyces tsukubaensis: tacrolimus (FK506) and butyrolactone synthetases production control. Appl Microbiol Biotechnol 98:4919–4936
Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y (1989) Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Bioeng 68:170–173
Schroeckh V, Scherlach K, Nützmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA (2009) Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106:14558–14563
Sheldon PJ, Busarow SB, Hutchinson CR (2002) Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol 44:449–460
Sidda JD, Poon V, Song L, Wang W, Yang K, Corre C (2016) Overproduction and identification of butyrolactones SCB1–8 in the antibiotic production superhost Streptomyces M1152. Org Biomol Chem 14:6390–6393
Stratigopoulos G, Cundliffe E (2002) Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem Biol 9:71–78
Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H (2010) Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci Biotechnol Biochem 74:819–827
Takano E (2006) γ-Butyrolactones: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9:287–294
Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ (1992) Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol Microbiol 6:2797–2804
Takano E, Chakaraburtty R, Nihira T, Yamada Y, Bibb MJ (2001) A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 41:1015–1028
Takano E, Kinoshita H, Mersinias V, Bucca G, Hotchkiss G, Nihira T, Smith CP, Bibb M, Wohlleben W, Chater K (2005) A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol 56:465–479
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333
Tang L, Grimm A, Zhang YX, Hutchinson CR (1996) Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol 22:801–813
Teshima A, Hadae N, Tsuda N, Arakawa K (2020) Functional analysis of P450 monooxygenase SrrO in the biosynthesis of butenolide-type signaling molecules in Streptomyces rochei. Biomolecules 10:1237
Thao NB, Kitani S, Nitta H, Tomioka T, Nihira T (2017) Discovering potential Streptomyces hormone producers by using disruptants of essential biosynthetic genes as indicator strains. J Antibiot 70:1004–1008
Vicente CM, Girardet JM, Hôtel L, Aigle B (2020) Molecular dynamics to elucidate the DNA-Binding activity of AlpZ, a member of the gamma-butyrolactone receptor family in Streptomyces ambofaciens. Front Microbiol 11:1255
Wang L, Vining LC (2003) Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW1, a member of the afsA family of gamma-butyrolactone regulatory genes. Microbiology 149:1991–2004
Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H (2009) Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A 106:8617–8622
Wang JB, Zhang F, Pu JY, Zhao J, Zhao QF, Tang GL (2014a) Characterization of AvaR1, an autoregulator receptor that negatively controls avermectins production in a high avermectin-producing strain. Biotechnol Lett 36:813–819
Wang W, Ji J, Wang J, Li S, Pan G, Fan K, Yang K (2014b) Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci U S A 111:5688–5693
Wang W, Zhang J, Liu X, Li D, Li Y, Tian Y, Tan H (2018) Identification of a butenolide signaling system that regulates nikkomycin biosynthesis in Streptomyces. J Biol Chem 293:20029–20040
Wietzorrek A, Bibb MJ (1997) A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184
Xu G, Yang S (2019) Regulatory and evolutionary roles of pseudo γ-butyrolactone receptors in antibiotic biosynthesis and resistance. Appl Microbiol Biotechnol 103:9373–9378
Xu G, Wang J, Wang L, Tian X, Yang H, Fan K, Yang K, Tan H (2010) “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem 285:27440–27448
Yamada Y (1995) Butyrolactone autoregulators, inducers of secondary metabolites, in Streptomyces. Actinomycetol 9:57–65
Yamamoto S, He Y, Arakawa K, Kinashi H (2008) γ-Butyrolactone-dependent expression of the SARP gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J Bacteriol 190:1308–1316
Yang K, Han L, Vining LC (1995) Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J Bacteriol 177:6111–6117
Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR (2010) A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol 400:847–864
Yu Q, Du A, Liu T, Deng Z, He X (2012) The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch Microbiol 194:415–426
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. ChemBioChem 10:625–633
Zhou S, Bhukya H, Malet N, Harrison PJ, Rea D, Belousoff MJ, Venugopal H, Sydor PK, Styles KM, Song L, Cryle MJ, Alkhalaf LM, Fülöp V, Challis GL, Corre C (2021) Molecular basis for control of antibiotic production by a bacterial hormone. Nature 590:463–467
Zhu J, Sun D, Liu W, Chen Z, Li J, Wen Y (2016) AvaR2, a pseudo γ-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol Microbiol 102:562–578
Zou Z, Du D, Zhang Y, Zhang J, Niu G, Tan H (2014) A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol Microbiol 94:490–505
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Arakawa, K., Suzuki, T. (2022). Regulation of Secondary Metabolites Through Signaling Molecules in Streptomyces. In: Rai, R.V., Bai, J.A. (eds) Natural Products from Actinomycetes. Springer, Singapore. https://doi.org/10.1007/978-981-16-6132-7_7
Download citation
DOI: https://doi.org/10.1007/978-981-16-6132-7_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-6131-0
Online ISBN: 978-981-16-6132-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)