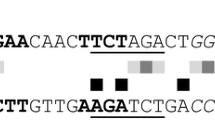
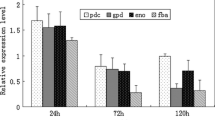
Abstract
Microbial xylanolytic enzymes have a promising biotechnological potential, and are extensively applied in industries. In this study, induction of xylanolytic activity was examined in Aspergillus phoenicis. Xylanase activity induced by xylan, xylose or β-methylxyloside was predominantly extracellular (93–97%). Addition of 1% glucose to media supplemented with xylan or xylose repressed xylanase production. Glucose repression was alleviated by addition of cAMP or dibutyryl-cAMP. These physiological observations were supported by a Northern analysis using part of the xylanase gene ApXLN as a probe. Gene transcription was shown to be induced by xylan, xylose, and β-methylxyloside, and was repressed by the addition of 1% glucose. Glucose repression was partially relieved by addition of cAMP or dibutyryl cAMP.



Similar content being viewed by others
References
Bhella RS, Altosaar I (1988) Role of cAMP in the mediation of glucose catabolite repression of glucoamylase synthesis in Aspergillus awamori. Curr Genet 14:247–252
Brühlmann F, Leupin M, Erismann KH, Fiechter A (2000) Enzymatic degumming of ramie bast fibers. J Biotechnol 76:43–50
Camacho NA, Aguilar OG (2003) Production, purification and characterization of a low molecular mass xylanases from Aspergillus sp. and its application in bakery. Appl Biochem Biotechnol 104:159–172
Collins T, Gerday C, Feller G (2005) Xylanases, xylanases families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23
Csiszár E, Urbánszki K, Szakás G (2001) Biotreatment of desized cotton fabric by commercial cellulase and xylanase enzymes. J Mol Catal B Enzym 11:1065–1072
Ghosh M, Nanda G (1994) Physiological studies on xylose induction and glucose repression of xylanolytic enzymes in Aspergillus sydowii MG49. FEBS Microbiol Lett 117:151–156
Graaff LH de, van den Broeck HC, Ooijen AJJ (1994) Regulation of the xylanase-encoding xlnA gene of Aspergillus tubigensis. Mol Microbiol 12:479–490
Hrmová M, Petraková E, Biely P (1991) Induction of cellulose and xylan-degrading enzyme system in Aspergillus terreus by homo and heterodisaccharides composed of glucose and xylose. J Gen Microbiol 137:541–547
Klich MA, Pitt JL (1998) A laboratory guide to common Aspergillus species and their telemorphs. Published by Commonwealth Scientific and Industrial Research Organization, Division of Food Processing
Kulkarni N, Shendye A, Rao M (1999) Molecular and biotechnological aspects of xylanases. FEMS Microbial Rev 23:411–456
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:267–275
Mach RL, Strauss J, Zeilinger S, Schindler M, Kubicek CP (1996) Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol Microbiol 21(6):1273–1281
Marui J, Tanaka A, Mimura S, de Graaff LH, Visser J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N (2002) A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet Biol 35:157–169
McIlvine TC (1921) A buffer solution for colorimetric comparison. J Biol Chem 49:183–186
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–429
Miyazaki K, Hirase T, Kojima Y, Flint HJ (2005) Medium- to large-sized xylo-oligosaccharides are responsible for xylanase induction in Prevotella bryantii B14. Microbiology 15:4121–4125
Morosoli R, Durand S, Boucher F (1989) Stimulation of xylanases synthesis in Cryptococcus albidus by cAMP. FEMS Microbiol Lett 57:57–60
Nakanishi K, Yasui T (1980) Kinetic studies on xylanase induction by β-xylosidase in Streptomyces sp. Agric Biol Chem 44:1885–1889
Ogasawara W, Shida Y, Furukawa T, Shimada R, Nakagawa S, Kawamura M, Yagyu T, Kosuge A, Xu J, Nogawa M, Okada H, Morikawa Y (2006) Cloning, functional expression and promoter analysis of xylanase III gene from Trichoderma reesei. Appl Microbiol Biotechnol 72:995–1003
Orejas M, MacCabe AP, Pérez-Gonzales JA, Kumar S, Ramón D (2001) The wide-domain carbon catabolite repressor CreA indirectly controls expression of the Aspergillus nidulans xlnB gene, encoding the acidic endo-β-(1,4)-xylanase X24. J Bacteriol 183(5):1517–23
Peij NNME van, Visser J, de Graaff LH (1998) Isolation and analysis of xlnR, encoding transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol 27:131–142
Piñaga F, Fernández-Espinar MT, Vallés S, Ramón D (1994) Xylanase production in Aspergillus nidulans, induction and carbon catabolite repression. FEMS Microbiol Lett 115:319–324
Polizeli MLTM, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS (2005) Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol 67:577–591
Prathumpai W, McIntyre M, Nielsen J (2004) The effect of CreA in glucose and catabolism in Aspergillus nidulans. Appl Microbiol Biotechnol 63:748–753
Raper KB, Fennell DI (1965) The genus Aspergillus. Williams & Wilkins, Baltimore
Ruijter GJG, Visser J (1997) Carbon repression in Aspergilli. FEMS Microbiol Lett 151:103–114
Saarelainen R, Paloheimo M, Fageström R, Suominen PL, Nevalainen KMH (1993) Cloning, sequencing and enhanced expression of the Trichoderma reesei endoxylanase II (pI 9) gene xln2. Mol Gen Genet 241:497–503
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbour Laboratory Press, Cold Spring Harbour
Sandrim VC, Rizzatti ACS, Terenzi HF, Jorge JA Milagres AMF, Polizeli MLTM (2005) Purification and biochemical characterization of two xylanases produced by Aspergillus caespitosus and their potential of kraft pulp bleaching. Process Biochem 40(5):1823–1828
Sokolovsky V, Kaldenhoff R, Ricci M, Russo VEA (1995) Fast and reliable mini-prep RNA extraction from Neurospora crassa. Fungal Genet Newsl 37:41–43
Strauss J, Mach RL, Zeilinger S, Stoffler G, Wolschek M, Hartler G, Kubicek CP (1995) Cre I the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett 376:103–107
Thevelein JM (1994) Signal transduction in yeast. Yeast 10:1753–1790
Törrönen A, Affenzeller KA, Hofer F, Myohanen TA, Blass D, Harkki A, Kubicek CP (1992) The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Biotechnology 10:1461–1465
Twomey LN, Pluske JR, Rowe JB, Choct M, Brown W, McConnell MF, Pethick DW (2003) The effects of increasing levels of soluble non-starch polysaccharides and inclusion of feeds enzymes in dog diets on faecal quality and digestibility. Anim Feed Sci Technol 108(1–4):71–82
Vogel HF (1964) Distribution of lysine pathways among fungi: evolutionary implications. Am Nat 98:435–446
Vries RP de, Visser J, de Graaff (1999) CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol 150:281–285
Weiland JJ (1997) Rapid procedure for the extraction of DNA from fungal spores and mycelia. Fungal Genet Newsl 44:60–63
Wong KKY, Tan LUL, Saddler JN (1998) Multiplicity of β-1,4-xylanase in microorganisms, functions and applications. Microbiol Rev 52(3):305–317
Xu J, Nogawa M, Okada H, Morikawa Y (2000) Regulation of xyn3 gene expression in Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol 54:370–375
Zonneveld BJM (1976) The effect of glucose and manganese on adenosine-3′,5′-monophosphate levels during growth and differentiation of Aspergillus nidulans. Arch Microbiol 108:41–44
Acknowledgments
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Conselho de Desenvolvimento Científico e Tecnológico (CNPq). M.C.B.; H.F.T.; J.A.J. and M.L.T.M.P. are Research Fellows of CNPq. A.C.S.R., F.Z.F. and S.C.P.N are Doctors recipient from CNPq. This work is part of A.C.S.R. thesis. The authors thank Ricardo Fernandes Alarcon, and Mauricio de Oliveira for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rizzatti, A.C.S., Freitas, F.Z., Bertolini, M.C. et al. Regulation of xylanase in Aspergillus phoenicis: a physiological and molecular approach. J Ind Microbiol Biotechnol 35, 237–244 (2008). https://doi.org/10.1007/s10295-007-0290-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-007-0290-9