Abstract
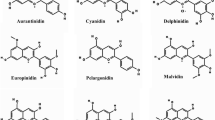
The enzymes flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) play an important role in flower color by determining the B-ring hydroxylation pattern of anthocyanins, the major floral pigments. F3′5′H is necessary for biosynthesis of the delphinidin-based anthocyanins that confer a violet or blue color to most plants. Antirrhinum majus does not produce delphinidin and lacks violet flower colour while A. kelloggii produces violet flowers containing delphinidin. To understand the cause of this inter-specific difference in the Antirrhinum genus, we isolated one F3′H and two F3′5′H homologues from the A. kelloggii petal cDNA library. Their amino acid sequences showed high identities to F3′Hs and F3′5′Hs of closely related species. Transgenic petunia expressing these genes had elevated amounts of cyanidin and delphinidin respectively, and flower color changes in the transgenics reflected the type of accumulated anthocyanidins. The results indicate that the homologs encode F3′H and F3′5′H, respectively, and that the ancestor of A. majus lost F3′5′H activity after its speciation from the ancestor of A. kelloggii.




Similar content being viewed by others
Abbreviations
- F3′H:
-
Flavonoid 3′-hydroxylase
- F3′5′H:
-
Flavonoid 3′,5′-hydroxylase
References
Brugliera F, Barri-Rewell G, Holton TA, Mason JG (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19:441–451
Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CaMV35S and MAS elements. Plant Mol Biol 15:373–381
Fukuchi-Mizutani M, Ohuhara H, Fukui Y, Nakao M, Katsumoto Y, Yonekura-Sakakibara K, Kusumi T, Hase T, Tanaka Y (2003) Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol 132:1652–1663
Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu C-Y, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366:276–279
Hoshino A, Morita Y, Choi JD, Saito N, Toki T, Tanaka Y, Iida S (2003) Spontaneous mutations of the flavonoid 3′-hydroxylase gene in the three morning glory species displaying reddish flowers. Plant Cell Physiol 44:990–1001
Jackson D, Roberts K, Martin C (1992) Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinurn majus. Plant J 2:425–434
Jones KN, Reithel JS (2001) Pollinator-mediated selection on a flower color polymorphism in experimental populations of Antirrhinum (Scrophulariaceae). Am J Bot 88:447–454
Katsumoto Y, Mizutani M, Fukui Y, Brugliera F, Holton T, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao G-Q, Nehra N, Lu C-Y, Dyson B, Tsuda S, Ashikari T, Kusumi T, Mason J, Tanaka Y (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48:1589–1600
Lazo G, Pascal A, Ludwig R (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. BioTechnology 9:963–967
Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinurn majus. Plant J 1:37–49
Nelson D, Werck-Reichhart D (2011) A P450-centric view of plant evolution. Plant J 66:194–211
Olmstead RG, DePamphilis CW, Wolfe AD, Young ND, Elisons WJ, Reeves PA (2001) Disintegration of the Scrophulariaceae. Am J Bot 88:348–361
Oyama RK, Baum DA (2004) Phylogenetic relationships of North American Antirrhinum (Veronicaceae). Am J Bot 91:918–925
Page RDM (1996) An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Rausher MD (2006) The evolution of flavonoids and their genes. In: Grotewold E (ed) The Science of flavonoids. Springer, Berlin, pp 175–211
Rausher MD (2008) Evolutionary transitions in floral color. Int J Plant Sci 169:7–21
Schwarz-Sommer Z, Davies B, Hudson A (2003) An everlasting pioneer: the story of Antirrhinum research. Nat Rev Genet 4:655–664
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18:831–851
Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G (2006) Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3,5′-hydroxylase cDNA reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol 61:365–381
Seitz C, Ameres S, Forkmann G (2007) Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett 581:3429–3434
Stickland RG, Harrison BJ (1974) Precursors and genetic control of pigmentation 1. Induced biosynthesis of pelargonidin, cyanidin and delphinidin in Antirrhinum majus. Heredity 33:108–112
Tanaka Y (2006) Flower colour and cytochromes P450. Phyochem Rev 5:283–291
Tanaka Y, Sasaki N, Ohmiya A (2008) Plant pigments for coloration: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Tanaka Y, Brugliera F, Chandler S (2009) Recent progress of flower colour modification by biotechnology. Int J Mol Sci 10:5350–5369
Ueyama U, Suzuki K, Fukuchi-Mizutani M, Fukui Y, Miyazaki K, Ohkawa H, Kusumi T, Tanaka Y (2002) Molecular and biochemical characterization of torenia flavonoid 3′-hydroxylase and flavone synthase II and modification of flower color by modulating the expression of these genes. Plant Sci 163:253–263
van Engelen FA, Molthoff JW, Conner AJ, Nap J, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4:288–290
Whibley AC, Langlade NB, Andalo C, Hanna AI, Bangham A, Thebaud C, Coen E (2006) Evolutionary paths underlying flower color variation in Antirrhinum. Science 313:963–966
Yoshida K, Mori M, Kondo T (2009) Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat Prod Rep 26:857–974
Zufall RA, Rausher MD (2003) The genetic basis of a flower color polymorphism in the common morning glory (Ipomoea purpurea). J Hered 94:442–448
Zufall RA, Rausher MD (2004) Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature 428:847–850
Acknowledgments
We thank Dr. Ludwig for providing Agrobacterium tumefaciens Agl0. Dr. Nelson is acknowledged for the nomenclature of A. kelloggii cytochromes P450. We are also grateful to Dr. Chandler for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ishiguro, K., Taniguchi, M. & Tanaka, Y. Functional analysis of Antirrhinum kelloggii flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes; critical role in flower color and evolution in the genus Antirrhinum . J Plant Res 125, 451–456 (2012). https://doi.org/10.1007/s10265-011-0455-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-011-0455-5