Abstract
Daphnia galeata is an important plankton in aquatic ecosystems. As a widely distributed species, D. galeata has been found throughout the Holarctic region. Understanding the genetic diversity and evolution of D. galeata requires the accumulation of genetic information from different locations. Even though the mitochondrial genome (mitogenome) sequence of D. galeata has already been reported, little is known about the evolution of its mitochondrial control region. In this study, D. galeata samples were collected from the Han River on the Korean Peninsula and its partial nd2 gene was sequenced for haplotype network analysis. This analysis showed that four clades of D. galeata were present in the Holarctic region. Moreover, the D. galeata examined in this study belonged to clade D and was specific to South Korea. The mitogenome of D. galeata from the Han River showed similar gene content and structure compared to sequences reported from Japan. Furthermore, the structure of control region of the Han River was similar to those of Japanese clones and differed substantially from European clone. Finally, a phylogenetic analysis based on the amino acid sequences of 13 protein-coding genes (PCGs) indicated that D. galeata from the Han River formed a cluster with clones collected from Lakes Kasumigaura, Shirakaba, and Kizaki in Japan. The differences in control region structure and stem and loop structure reflect the different evolutionary directions of the mitogenomes from Asian and European clones. These findings improve our understanding of the mitogenome structure and genetic diversity of D. galeata.




Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in Genbank under the accession number OM397534.
References
Athanasio CG, Chipman JK, Viant MR, Mirbahai L (2016) Optimisation of DNA extraction from the crustacean Daphnia. PeerJ 10:4–e2004. https://doi.org/10.7717/peerj.2004
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Bernatowicz P, Dawidowicz P, Pijanowska J (2021) Phenotypic plasticity and developmental noise in hybrid and parental clones of Daphnia longispina complex. Aquat Ecol 55:1179–1188. https://doi.org/10.1007/s10452-021-09898-7
Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phyl Evol 69:313–319. https://doi.org/10.1016/j.ympev.2012.08.023
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780. https://doi.org/10.1093/nar/27.8.1767
Brooks JL (1957) The systematics of North American Daphnia. Mem Conn Acad Arts Sci 13:1–180
Brown WM, George M, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci 76:1967–1971. https://doi.org/10.1073/pnas.76.4.196
Campbell NJH, Sturm RA, Barker SC (2001) Large mitochondrial repeats multiplied during the polymerase chain reaction. Mol Ecol Notes 1:336–340. https://doi.org/10.1046/j.1471-8278.2001.00093.x
Chao QJ, Li YD, Geng XX, Zhang L, Dai X, Zhang X, Li J, Zhang HJ (2014) Complete mitochondrial genome sequence of Marmota himalayana (Rodentia: Sciuridae) and phylogenetic analysis within Rodentia. Genet Mol 13:2739–2751. https://doi.org/10.4238/2014.April.14.3
Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705. https://doi.org/10.1016/0092-8674(82)90049-6
Crease TJ (1999) The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea). Gene 233:89–99. https://doi.org/10.1016/S0378-1119(99)00151-1
Edmondson WT, Litt AH (1982) Daphnia in Lake Washington. Limnol Oceanogr 27:272–293. https://doi.org/10.4319/lo.1982.27.2.0272
Fields PD, McTaggart S, Reisser CM, Haag C, Palmer WH, Little TJ, Ebert D, Obbard DJ (2022) Population-genomic analysis identifies a low rate of global adaptive fixation in the proteins of the cyclical parthenogen Daphnia magna. Mol Biol Evol 39:msac048. https://doi.org/10.1093/molbev/msac048
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase I from diverse metazoan invertebrates. Mol Marina Biol Technol 3(5):294–299
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res 41:e129. https://doi.org/10.1093/nar/gkt371
Ishida S, Taylor DJ (2007) Quaternary diversification in a sexual Holarctic zooplankter, Daphnia galeata. Mol Ecol 16:569–582. https://doi.org/10.1111/j.1365-294X.2006.03160.x
Ishida S, Kotov AA, Taylor DJ (2006) A new divergent lineage of Daphnia (Cladocera: Anomopoda) and its morphological and genetical differentiation from Daphnia curvirostris Eylmann, 1887. Zool J Linn Soc 146:385–405. https://doi.org/10.1111/j.1096-3642.2006.00214.x
Jiang XZ, Du NS (1979) Fauna sinica: Crustacean-freshwater Cladocera. Science Press, pp 102–118
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kuhn K, Streit B, Schwenk K (2008) Conservation of structural elements in the mitochondrial control region of Daphnia. Gene 420:107–112. https://doi.org/10.1016/j.gene.2008.05.020
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Laslett D, Canback B (2007) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24:172–175. https://doi.org/10.1093/bioinformatics/btm573
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221. https://doi.org/10.1093/oxfordjournals.molbev.a040442
Lin ZJ, Wang X, Wang J, Tan Y, Tang X, Werren JH, Zhang D, Wang X (2021) Comparative analysis reveals the expansion of mitochondrial DNA control region containing unusually high G-C tandem repeat arrays in Nasonia vitripennis. Int J Biol Macromol 166:1246–1257. https://doi.org/10.1016/j.ijbiomac.2020.11.007
Liu Q, Deng D, Zhang K, He P, Sun Y, Zhang T, Yang W, Liu W (2019) Genetic diversity and differentiation of Daphnia galeata in the middle and lower reaches of the Yangtze River, China. Ecol Evol 9:12688–12700. https://doi.org/10.1002/ece3.5737
Lu Y, Johnston PR, Dennis SR, Monaghan MT, John U, Spaak P, Wolinska J (2018) Daphnia galeata responds to the exposure to an ichthyosporean gut parasite by down-regulation of immunity and lipid metabolism. BMC Genomics 19:932. https://doi.org/10.1186/s12864-018-5312-7
Lunt DH, Whipple LE, Hyman BC (1998) Mitochondrial DNA variable number tandem repeats (VNTRs): utility and problems in molecular ecology. Mol Ecol 7:1441–1455. https://doi.org/10.1046/j.1365-294x.1998.00495.x
Ma Y, He K, Yu P, Yu D, Cheng X, Zhang J (2015) The complete mitochondrial genomes of three bristletails (Insecta: Archaeognatha): the paraphyly of Machilidae and insights into archaeognathan phylogeny. PLoS One 10:e0117669. https://doi.org/10.1371/journal.pone.0117669
Nickel J, Schell T, Holtzem T, Thielsch A, Dennis SR, Schlick-Steiner BC, Steiner FM, Most M, Pfenninger M, Schwenk K (2021) Hybridization dynamics and extensive introgression in the Daphnia longispina species complex: new insights from a high-quality Daphnia galeata reference genome. Genome Biol Evol 13:1–23. https://doi.org/10.1093/gbe/evab267
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474. https://doi.org/10.1038/290470a0
Ozdemir E, Altındag A, Kandemir I (2016) Molecular diversity of some species belonging to the genus Daphnia O. F. Müller, 1785 (Crustacea: Cladocera) in Turkey. Mitochondrial DNA A DNA Mapp Seq Anal 28:424–433. https://doi.org/10.3109/19401736.2015.1136303
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358. https://doi.org/10.1007/BF00186547
Polakova SB, Lichtner Z, Szemes T, Smolejova M, Sulo P (2021) Mitochondrial DNA duplication, recombination, and introgression during interspecific hybridization. Sci Rep 11:12726. https://doi.org/10.1038/s41598-021-92125-y
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 12:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Shadel GS, Clayton DA (1993) Mitochondrial transcription initiation – variation and conservation. J Biol Chem 268:16083–16086. https://doi.org/10.1016/S0021-9258(19)85387-5
Shapiro J (1980) The importance of trophic-level interactions to the abundance and species composition of algae in lakes. Hypertrophic Ecosystems 2:105–116. https://doi.org/10.1007/978-94-009-9203-0_12
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of parge phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. https://doi.org/10.1080/10635150701472164
Tams V, Nickel JH, Ehring A, Cordellier M (2020) Insights into the genetic basis of predator-induced response in Daphnia galeata. Ecol Evol 10:13095–13108. https://doi.org/10.1002/ece3.6899
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Tokishita SI, Shibuya H, Kobayashi T, Sakamoto M, Ha JY, Yokobori SI, Yamagata H, Hanazato T (2017) Diversification of mitochondrial genome of Daphnia galeata (Cladocera, Crustacea): comparison with phylogenetic consideration of the complete sequences of clones isolated from five lakes in Japan. Gene 611:38–46. https://doi.org/10.1016/j.gene.2017.02.019
Turner DH, Mathews DH (2010) NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38(Database issue):D280–D282. https://doi.org/10.1093/nar/gkp892
Vo TMC, Pham NH, Nguyen TD, Bui MH, Dao TS (2018) Development of Daphnia magna under exposure to ampicillin. Architec. Civil Engin Environ 11:147–152. https://doi.org/10.21307/ACEE-2018-047
Watanabe Y, Suematsu T, Ohtsuki T (2014) Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front Genet 5:109. https://doi.org/10.3389/fgene.2014.00109
Wei W, Giebler S, Wolinska J, Ma X, Yang Z, Hu W, Yin M (2015) Genetic structure of Daphnia galeata populations in Eastern China. PLoS One 10:e0120168. https://doi.org/10.1371/journal.pone.0120168
Wilson K, Cahill V, Ballment E, Benzie J (2000) The complete sequence of the mitochondrial genome of the crustacean Penaeus monodon: are malacostracan crustaceans more closely related to insects than to branchiopods? Mol Biol 17:863–874. https://doi.org/10.1093/oxfordjournals.molbev.a026366
Xu L, Lin Q, Xu S, Gu Y, Hou J, Liu Y, Dumont HJ, Han BP (2018) Daphnia diversity on the Tibetan Plateau measured by DNA taxonomy. Ecol Evol 8:5069–5078. https://doi.org/10.1002/ece3.4071
Zhang DX, Hewitt GM (1997) Insect mitochondrial control region: a review of its structure, evolutionary studies. Biochem Syst Ecol 25:99–120. https://doi.org/10.1016/S0305-1978(96)00042-7
Zhang DX, Szymura JM, Hewitt GM (1995) Evolution and structural conservation of the control region of insect mitochondrial-DNA. J Mol Evol 40:382–391. https://doi.org/10.1007/BF00164024
Lampert W (2011) Daphnia: development of a model organism in ecology and evolution. Excell Ecol 21:1–275. Inter Ecol Ins
Rambaut A (2014) FigTree v1.4.4. Available at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 February 2022)
Acknowledgements
We acknowledged Jong-Won Baek and Min-Ho Mun (Sangmyung University, South Korea) sample collection for the assistance of sample collection in field.
Funding
This work was supported by the National Research Foundation of South Korea (NRF) grant funded by the South Korea government (MSIP) (No. NRF-2016M3A9E1915578).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by CBK, TJC, TDD, and JIK. The first draft of the manuscript was written by TJC and TDD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1:
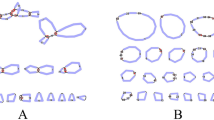
Table S1 nd2 sequences of Daphnia galeata obtained for haplotype network analysis from Genbank used in this study. Table S2 Similarity of orthologous alpha/alpha’ and gamma/gamma’ of Daphnia galeata.Fig. S1 Median-Joining network of haplotypes based on the variation of the mitochondrial nd2 gene observed in the Holarctic populations of Daphnia galeata, Blue indicates the Han River clone analyzed in this study. Mutated positions are shown as branch lines.Fig. S2 Transfer RNA secondary structures for Daphnia galeata collected from the Han River, South Korea.Fig. S3 Phylogenetic relationships of Daphnia galeata based on amino acid sequences of 13 PCGs. Black circle indicates the newly sequenced mitogenome in this study. GenBank accession numbers are indicated next to species names. Possibility values (left) and bootstrap values (right) are shown at the nodes. (DOCX 327 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, TJ., Do, T.D., Kim, JI. et al. Analysis of the complete mitogenome of Daphnia galeata from the Han River, South Korea: structure comparison and control region evolution. Funct Integr Genomics 23, 65 (2023). https://doi.org/10.1007/s10142-023-00986-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-00986-5