Abstract
Extensive transcriptomic skimming was conducted to decipher molecular, morphological, physiological, and biochemical responses in salt-tolerant (PDL-1) and salt-sensitive (L-4076) cultivars under control (0 mM NaCl) and salinity stress (120 mM NaCl) conditions at seedling stage. Morphological, physiological, and biochemical studies revealed that PDL-1 exhibited no salt injury and had higher K+/Na+ ratio, relative water content (RWC), chlorophyll, glycine betaine, and soluble sugars in leaves while lower H2O2 induced fluorescence signals in roots as compared to L-4076. Transcriptomic profile revealed a total of 17,433 significant differentially expressed genes (DEGs) under different treatments and cultivar combinations that include 2557 upregulated and 1533 downregulated transcripts between contrasting cultivars under salt stress. Accuracy of transcriptomic analysis was validated through quantification of 10 DEGs via quantitative real-time polymerase chain reaction (qRT-PCR). DEGs were functionally characterized by Gene Ontology (GO) analysis and assigned to various metabolic pathways using MapMan. DEGs were found to be significantly associated with phytohormone-mediated signal transduction, cellular redox homoeostasis, secondary metabolism, nitrogen metabolism, and cellular stress signaling. The present study revealed putative molecular mechanism of salinity tolerance in lentil together with identification of 5643 simple sequence repeats (SSRs) and 176,433 single nucleotide polymorphisms (SNPs) which can be utilized to enhance linkage maps density along with detection of quantitative trait loci (QTLs) associated with traits of interests. Stress-related pathways identified in this study divulged plant functioning that can be targeted to improve salinity stress tolerance in crop species.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. Sequences were deposited in Sequence Read Archive (SRA) (BioProject Accession: PRJNA423135).
References
Abdelgadir EM, M Oka, H Fujiyama (2005) Characteristics of nitrate uptake by plants under salinity. J Plant Nutr 28:33–46. https://doi.org/10.1081/PLN-200042156
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78. https://doi.org/10.1105/tpc006130
Ahmad A, Ismun A, Taib M, Ilias MA, Ismail A, Othman R, Zainudin SR (2017) Effects of salinity stress on carbohydrate metabolism in Cryptocoryne elliptica cultures. J Trop Plant Physiol 9:1–13
Ahmed S, Choudhury AR, Chatterjee P, Samaddar S, Kim K, Jeon S, Sa T (2019) The role of plant growth-promoting rhizobacteria to modulate proline biosynthesis in plants for salt stress alleviation. In: Sayyed RZ, Arora NK, Reddy MS (eds) Plant growth promoting rhizobacteria for sustainable stress management. Springer, Singapore, pp 1–20
Almeida DM, Oliveira MM, Saibo NJ (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Gen Mol Bio 40(1):326-345. https://doi.org/10.1590/1678-4685-gmb-2016-0106
An JP, Yao JF, Xu RR, You CX, Wang XF, Hao YJ (2018) An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol Plant1 64(3):279–289. https://doi.org/10.1111/ppl12724
Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59(2):206–216. https://doi.org/10.1016/j.envexpbot.2005.12.006
Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166(1):3–16. https://doi.org/10.1016/j.plantsci.2003.10.024
Aswath CR, Kim SH, Mo SY, Kim DH (2005) Transgenic plants of creeping bent grass harboring the stress inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl stress. J Plant Growth Regul 47(2-3):129-139. https://doi.org/10.3389/fphys201700509
Auclair SM, Bhanu MK, Kendall DA (2012) Signal peptidase I: cleaving the way to mature proteins. Protein Sci 21(1): 13-25. https://doi.org/10.1002/pro757
Azooz MM (2009) Salt stress mitigation by seed priming with salicylic acid in two faba bean genotypes differing in salt tolerance. Int J Agric Biol 11(4):343–350
Bandeoglu E, Eyidogan F, Yucel M, Oktem HA (2004) Antioxidant responses of shoots and roots of lentil to NaCl-salinity stress. Plant Growth Regul 42(1):69-77. https://doi.org/10.1023/B:GROW0000014891354277b
Bertram R, Pedersen MG, Luciani DS, Sherman A (2006) A simplified model for mitochondrial ATP production. J Theor Biol 243(4):575-586. https://doi.org/10.1016/jjtbi200607019
Bhatla S C (2018) Jasmonic acid. In: Plant physiology, development and metabolism. Springer pp. 671–679
Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12(4):431-434. https://doi.org/10.1016/s0955-0674(00)00112-5
Bolger AM, Lohse M,Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data Bioinformatics30(15):2114-2120. https://doi.org/10.1093/bioinformatics/btu170
Boursiac Y, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Maurel C (2008) Stimulus-induced down-regulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J56 (2):207-218. https://doi.org/10.1111/j1365-313x200803594x
Brankova L, Ivanov S, Alexieva V (2007) The induction of microsomal NADPH: cytochrome P450 and NADH: cytochrome b5 reductases by long-term salt treatment of cotton (Gossypium hirsutum L) and bean (Phaseolus vulgaris L) plants. Plant Physiol Biochem 45(9):691-695. https://doi.org/10.1016/jplaphy200707005
Cai X, Zhang C, Shu W, Ye Z, Li H, Zhang Y (2016) The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem Biophys Res Commun 474(4):736–741
Cao D, Hou W, Liu W, Yao W, Wu C, Liu X (2011) Overexpression of TaNHX2 enhances salt tolerance of “composite” and whole transgenic soybean plants. Plant Cell Tissue Organ Cult 107:541–552. https://doi.org/10.1007/s11240-011-0005-9
Carillo P, Annunziata MG, Pontecorvo G, Fuggi A,Woodrow P (2011) Salinity stress and salt tolerance. In: Shanker A, Arun, Venkateshwarlu B (eds) Abiotic stress in plants: mechanisms and adaptationspp 21-38
Caverzan A, Casassola A, Brammer S P (2016)Antioxidant responses of wheat plants under stress. Genet Mol Biol 39(1):1-6. https://doi.org/10.1590/1678-4685-GMB-2015-0109
Chakraborty K, Sairam RK, Bhattacharya RC (2012) Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol Biochem51:90–101 https://doi.org/10.1016/jplaphy201110001
Chen H, An R, Tang JH, Cui XH, Hao, FS, Chen J, Wang XC (2007) Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Mol Breed 19(3):215–225 https://doi.org/10.1007/s11032-006-9048-8
Cherel I (2004) Regulation of K+ channel activities in plants: from physiological to molecular aspects. J Exp Bot55:337–51. https://doi.org/10.1093/jxb/erh028
Conde A, Chaves MM, Gerós H (2011)Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol 52(9):1583-1602. https://doi.org/10.1093/pcp/pcr107
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674-3676. https://doi.org/10.1093/bioinformatics/bti610
Datir S, Singh N, Joshi I (2020) Effect of NaCl-induced salinity stress on growth, osmolytes and enzyme activities in wheat genotypes. Bull Environ Contam Toxicol104(3):351-357. https://doi.org/10.1007/s00128-020-02795-z
Demiİr Y, Kocaçalişkan I (2001) Effects of NaCl and proline on polyphenol oxidase activity in bean seedlings. Biol Plant 44(4):607-609. https://doi.org/10.1023/A:1013715425310
Denancé N, Ranocha P, Oria N, Barlet X, Rivière MP, Yadeta KA, Hoffmann L, Perreau F, Clément G, Maia-Grondard A, Van den Berg GC (2013) Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J 73(2):225–239
Dong W, Ai X, Xu F, Quan T, Liu S, Xia G (2012) Isolation and characterization of a bread wheat salinity responsive ERF transcription factor. Gene 511(1):38–45. https://doi.org/10.1016/jgene201209039
Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan, Guy C, Smith SM, Steup M, Ritte G (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial β-amylases. Plant Physiol 145(1):17–28
Evers D, Legay S, Lamoureux D, Hausman JF, Hoffmann L, Renaut J (2012)Towards a synthetic view of potato cold and salt stress response by transcriptomic and proteomic analyses. Plant Mol Biol 78(4-5):503-514. https://doi.org/10.1007/s11103-012-9879-0
Fahad S, Hussain S, Matloob A, Khan FA, Khaliq A, Saud S, Hassan S, Shan D, Khan F, Ullah N, Faiq M (2015) Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul 75(2): 391-404. https://doi.org/10.1007/s10725-014-0013-y
Farooq M, Gogoi N, Hussain M, Barthakur S, Paul S Bharadwaj N, Migdadi HM, Alghamdi SS, Siddique KH (2017) Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol Biochem 118:199-217. https://doi.org/10.1016/jplaphy201706020
Fini A, Brunetti C, Di Ferdinando M, Ferrini F, Tattini M (2011)Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav 6(5):709-711. https://doi.org/10.4161/psb6515069
Foti C, Khah EM, Pavli OI (2019) Germination profiling of lentil genotypes subjected to salinity stress. Plant Biol 21(3):480-486. https://doi.org/10.1111/plb12714
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45(2):146-159. https://doi.org/10.1093/pcp/pch014
Garg R, Chevala VN, Shankar R, Jain M(2015) Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci Rep 5:14922. https://doi.org/10.1038/srep14922
Garg R, Verma M, Agrawa LS, Shankar R, Majee M, Jain M (2014) Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel Insights into the salinity and submergence tolerance factors. DNA Res 21:69–84. https://doi.org/10.1093/dnares/dst042
Gigon A, Matos AR, Laffray D, Zuily-Fodil Y, Pham-Thi AT (2004) Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann Bot 94(3):345-351. https://doi.org/10.1093/aob/mch150
Giri J (2011) Glycine betaine and abiotic stress tolerance in plants. Plant Signal Behav 6(11):1746-1751 https://doi.org/10.1093/aob/mch150
Golldack D, Lüking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30(8):1383-1391. https://doi.org/10.1007/s00299-011-1068-0
Gonzalez DH (2016) Introduction to transcription factor structure and function. In: Meshi T, Iwabuchi M (eds) Plant transcription factors. Academic Press, pp 3–11
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Chen Z (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644. https://doi.org/10.1038/nbt1883
Guo Y, Huang C, Xie Y, Song F,Zhou X (2010) A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 232(6):1499-1509. https://doi.org/10.1007/s00425-010-1271-1
Hadi MR, Karimi N (2012) The role of calcium in plants salt tolerance. J Plant Nutr 35(13):2037–2054. https://doi.org/10.1080/019041672012717158
Handy DE, Loscalzo J (2012) Redox regulation of mitochondrial function. Antioxid Redox Sign 16(11):1323-1367. https://doi.org/10.1016/jredox201312011
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499. https://doi.org/10.1146/annurev.arplant.51.1.463
Hertel S, Zoschke R, Neumann L, Qu Y, Axmann I M, Schmitz-Linneweber C (2013) Multiple checkpoints for the expression of the chloroplast-encoded splicing factor Mat K. Plant Physiol 163(4):1686-1698. https://doi.org/10.1104/pp113227579
Hiz MC, Canher B, Niron H, Turet M (2014) Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L) under saline conditions PloS ONE, 9(3) https://doi.org/10.1371/journalpone0092598
Hossain MS, Dietz KJ (2016) Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front Plant Sci 7:548. https://doi.org/10.3389/fpls201600548
Hossain MA, Uddin MK, Ismail MR, Ashrafuzzaman M (2012) Responses of glutamine synthetase-glutamate synthase cycle enzymes in tomato leaves under salinity stress. Int J Agric Biol 14(4):509–515
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67(1-2):69-181. https://doi.org/10.1007/s11103-008-9309-5
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23(15):1805-1817 https://doi.org/10.1101/gad1812409
Jacoby RP, Taylor NL, Millar AH (2011) The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci 16(11):614-623. https://doi.org/10.1016/jtplants201108002
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6(2):275-286. https://doi.org/10.1093/mp/sst017
Jiang SC, Mei C, Liang S, Yu YT, Lu K, Wu Z, Zhang DP (2015) Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol Biol 88(4-5):369-385. https://doi.org/10.1007/s11103-015-0327-9
Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Ronald PC (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152(3):1674-1692. https://doi.org/10.1104/pp109152157
Kabbage M, Dickman, MB (2008) The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci 65(9):1390–1402. https://doi.org/10.1007/s00018-008-7535-2
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PloS ONE 7(6). https://doi.org/10.1371/journalpone0040203
Kawakami EM, Oosterhuis DM, Snider JL (2013) Nitrogen assimilation and growth of cotton seedlings under NaCl salinity and in response to urea application with NBPT and DCD. J Agron Crop Sci 199(2):106-117 https://doi.org/10.1111/jac12002
Khan MI, Asgher M, Khan NA (2014) Alleviation of salt-induced photosynthesis and growth inhibition by salicylic acid involves glycine betaine and ethylene in mungbean (Vigna radiata L.) Plant Physiol Biochem 80:67–74
Koyro HW (2006) Effect of salinity on growth, photosynthesis, water relations and solute composition of the potential cash crop halophyte Plantagocoronopus (L). Environ Exp Bot 56(2):136-146. https://doi.org/10.1016/jenvexpbot200502001
Krishnamurthy P, Vishal B, Khoo K, Rajappa S, Loh CS, Kumar PP (2019) Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis. Plant Cell Rep 38(10):1299-1315. https://doi.org/10.1007/s00299-019-02450
Laffray X, Alaoui-Sehmer L, Bourioug M, Bourgeade P, Alaoui-Sossé B, Aleya L (2018) Effects of sodium chloride salinity on ecophysiological and biochemical parameters of oak seedlings (Quercus robur L) from use of de-icing salts for winter road maintenance Environ Monit Assess 190(5): 266 https://doi.org/10.1007/s10661-018-6645-z
Langmead, Schatz MC, Lin J, Pop M, Salzberg SL (2009) Searching for SNPs with cloud computing. Genome Biol 10(11):1–10. https://doi.org/10.1186/gb-2009-10-11-r134
Lim CW, Han SW, Hwang IS, Kim DS, Hwang BK, Lee SC (2015) The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol 56(5):930–42. https://doi.org/10.1093/pcp/pcv020
Machado RMA, Serralheiro RP (2017) Soil salinity: effect on vegetable crop growth management practices to prevent and mitigate soil salinization. Horticulturae 3(2):30 https://doi.org/10.3390/horticulturae3020030
Manishankar P, Wang N, Köster P, Alatar AA, Kudla J (2018) Calcium signaling during salt stress and in the regulation of ion homeostasis. J Exp Bot 69(17): 4215-4226. https://doi.org/10.1093/jxb/ery201
Masuda Y, Kamisaka S, Yanagisawa H, Suzuki Y (1981) Effect of light on growth and metabolic activities in pea seedlings I changes in cell wall polysaccharides during growth in the dark and in the light. Biochem Physiol Pflanz 176(1):23-34. https://doi.org/10.1016/S0015-3796(81)80005-4
Mehta PA, Sivaprakash K, Parani M, Venkataraman G, Parida AK (2005) Generation and analysis of expressed sequence tags from the salt-tolerant mangrove species Avicennia marina (Forsk) Vierh. Theor Appl Genet 110(3):416–424. https://doi.org/10.1007/s00122-004-1801-y
Mialoundama AS, Jadid N, Brunel J, Di Pascoli T, Heintz D, Erhardt M, RahierA (2013) Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. The Plant Cell 25(12):4879–4893. https://doi.org/10.1105/tpc113115576
Mian A,Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, Véry AA (2011) Overexpression of a Na+ and K+ -permeable HKT transporter in barley improves salt tolerance. Plant J. 68: 468–479. https://doi.org/10.3390/genes9100475
Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy MK (2007) Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol Biol 64:713–732. https://doi.org/10.1007/s11103-007-9193-4
Misra N, Saxena P (2009) Effect of salicylic acid on proline metabolism in lentil grown under salinity stress. Plant Sci. 177(3):181–189. https://doi.org/10.1016/jplantsci200905007
Muthukumarasamy M, Gupta SD, Panneerselvam R (2000) Enhancement of peroxidase, polyphenol oxidase and superoxide dismutase activities by triadimefon in NaCl stressed Raphanus sativus L. Biol Plant 43(2): 317-320. https://doi.org/10.1023/A:1002741302485
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Bba-Gene Regul Mech 1819(2):97-103. https://doi.org/10.1016/jbbagrm201110005
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34(2):137-148. https://doi.org/10.1046/j1365-313X200301708x
Ning X, Sun Y, Wang C, Zhang W, Sun M, Hu H, Yang L (2018) A rice CPYC-type glutaredoxin OsGRX20 in protection against bacterial blight, methyl viologen and salt stresses. Front Plant Sci9:111. https://doi.org/10.3389/fpls201800111
Palma F, López-Gómez M, Tejera NA, Lluch C (2013) Salicylic acid improves the salinity tolerance of Medicago sativa in symbiosis with Sinorhizobium meliloti by preventing nitrogen fixation inhibition. Plant Sci 208:75–82
Pao GM, Wu LF, Johnson KD, Höfte H, Chrispeels MJ, Sweet G, Saier Jr MH (1991) Evolution of the MIP family of integral membrane transport proteins. Mol Microbiol 5(1):33-37 https://doi.org/10.1111/j1365-29581991tb01823x
Pardo JM, Quintero FJ (2002) Plants and sodium ions: keeping company with the enemy. Genome Biol 3(6):reviews1017. https://doi.org/10.1186/gb-2002-3-6-reviews1017
Passricha N, Saifi SK, Kharb P, Tuteja N (2019) Marker-free transgenic rice plant overexpressing pea LecRLK imparts salinity tolerance by inhibiting sodium accumulation. Plant Mol Biol 99(3):265-281. https://doi.org/10.1007/s11103-018-0816-8
Popova LP, Stoinova ZG, Maslenkova LT (1995) Involvement of abscisic acid in photosynthetic process in Hordeum vulgare L. during salinity stress. Plant Growth Regul 14(4):211.https://doi.org/10.1007/BF00204914
Pratap A, Kumar J, Kumar S (2014) Evaluation of wild species of lentil for agro-morphological traits. Legume Res 37(1):11–18 https://hdl.handle.net/2050011766/7290
Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol 129(1):145-155. https://doi.org/10.1104/pp010988
Rienth M, Torregrosa L, Luchaire N, Chatbanyong R, Lecourieux D, Kelly MT, Romieu C (2014) Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol 14(1): 1-18. https://doi.org/10.1186/1471-2229-14-108
Sairam RK, Rao KV, Srivastava GC (2002) Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci 163(5):1037-1046. https://doi.org/10.1016/S0168-9452(02)00278-9
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133(4):651-669. https://doi.org/10.1111/j1399-3054200701008x
Shavrukov Y (2013) Salt stress or salt shock: which genes are we studying? J Exp Bot 64(1):119-127. https://doi.org/10.1093/jxb/ers316
Shi H, Quintero FJ, Pardo JM,Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14(2):465-477. https://doi.org/10.1105/tpc010371
Shore D, Bianchi A (2009) Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J 28(16):2309-2322. https://doi.org/10.1038/emboj2009195
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation.Saudi J Biol Sci 22(2):123–131 https://doi.org/10.1016/jsjbs201412001
Sidari M, Santonoceto C, Anastasi U, Preiti G, Muscolo A (2008) Variations in four genotypes of lentil under NaCl-salinity stress. J Agric Sci 3:410-416. https://doi.org/10.3844/ajabssp2008410416
Singh D, Pal M, Singh CK, Taunk J, Jain P, Chaturvedi AK, Nongthombam R (2016b) Molecular scanning and morpho-physiological dissection of component mechanism in Lens species in response to aluminium stress. PloS ONE, 11(7): e0160073. https://doi.org/10.1371/journalpone0160073
Singh D, Singh CK, Kumari S, Tomar RSS, Karwa S, Singh R, Pal M (2017a) Discerning morpho-anatomical, physiological and molecular multiformity in cultivated and wild genotypes of lentil with reconciliation to salinity stress. PloS ONE, 12(5). https://doi.org/10.1371/journalpone0177465
Singh D, Singh CK, Taunk J, Jadon V, Pal M, Gaikwad K (2019) Genome wide transcriptome analysis reveals vital role of heat responsive genes in regulatory mechanisms of lentil (Lens culinaris Medikus). Sci Rep 9(1):1-19. https://doi.org/10.1038/s41598-019-49496-0
Singh D, Singh C K, Taunk J, Tomar RSS, Chaturvedi AK, Gaikwad K, Pal M (2017b) Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genomics 18(1):206. https://doi.org/10.1186/s12864-017-3596-7
Singh D, Singh CK, Tomar RSS, Taunk J, Singh R, Maurya S, Kumar DS (2016a) Molecular assortment of Lens species with different adaptations to drought conditions using SSR markers. PloS ONE 11(1) https://doi.org/10.1371/journalpone0147213
Song S Y, Chen Y, Chen J, Dai, XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234(2):331–345 https://doi.org/10.1007/s00425-011-1403-2
Sreenivasulu N, Ramanjulu S, Ramachandra-Kini K, Prakash H S, Shekar-Shetty H, Savithri H S, Sudhakar C (1999) Total peroxidase activity and peroxidase isoforms as modified by salt stress in two cultivars of fox-tail millet with differential salt tolerance. Plant Sci 141(1): 1-9. https://doi.org/10.1016/S0168-9452(98)00204-0
Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48(8):1148-1158. https://doi.org/10.1093/pcp/pcm088
Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Aida M (2011) CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J 66(6):1066-1077. https://doi.org/10.1111/j1365-313X201104571x
Tan T, Cai J, Zhan E, Yang Y, Zhao J, Guo Y, Zhou H (2016) Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol Biol 92(3):391–400
Tenhaken R (2015) Cell wall remodeling under abiotic stress. Front Plant Sci 5:771. https://doi.org/10.3389/fpls.2014.00771
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants Ann Bot 91(5):503-527. https://doi.org/10.1093/aob/mcg058
Tsonev TD, Lazova GN, Stoinova ZG, Popova LP (1998) Apossible role for Jasmonic acid in adaptation of barley seedlings to salinity stress. J Plant Growth Regul 17:153–159. https://doi.org/10.1007/PL00007029
Tuteja R (2005) Type I signal peptidase: an overview. Arch Biochem Biophys 441(2):107-111. https://doi.org/10.1016/jabb200507013
Wang YW, Chen SM, Wang WJ, Huang XQ, Zhou CF, Zhuang Z,Lu S (2016) The DnaJ-like zinc finger domain protein PSA2 affects light acclimation and chloroplast development in Arabidopsis thaliana. Front Plant Sci 7:360. https://doi.org/10.3389/fpls201600360
Wang F, Jing W, Zhang W (2014) The mitogen-activated protein kinase cascade MKK1–MPK4 mediates salt signaling in rice. Plant Sci 227:181-189. https://doi.org/10.1016/jplantsci201408007
Wang Y, Li K, Li X (2009) Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J Plant Physiol 166(15):1637-1645. https://doi.org/10.1016/jjplph200904009
Wang J, Xu M, Gu Y, Xu LA (2017) Differentially expressed gene analysis of Tamarix chinensis provides insights into NaCl-stress response. Trees 31(2):645–658. https://doi.org/10.1007/s00468-016-1497-z
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci10:228. https://doi.org/10.3389/fpls201900228
Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM (2004) Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167(4):849-859. https://doi.org/10.1016/jplantsci200405034
Yamaguchi M, Demura T (2010) Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol 27(3):237–242. https://doi.org/10.5511/plantbiotechnology.27.237
Yang Y, Guo Y (2018) Unraveling salt stress signaling in plants. J Integr Plant Biol 60(9):796-804. https://doi.org/10.1111/jipb12689
Yang S, Zhao L, Yan J, Zhang J, Guo F, Geng Y, Li X (2019) Peanut genes encoding tetrapyrrole biosynthetic enzymes, AhHEMA1 and AhFC1, alleviating the salt stress in transgenic tobacco. Plant Physiol Biochem 137:14–24. https://doi.org/10.5511/plantbiotechnology.27.237
Yano R, Nakamura M, Yoneyama T, Nishida I (2005) Starch-related α-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol 138(2):837-846.https://doi.org/10.1104/pp104056374
Yu Y, T Xu, X Li, J Tang, D Ma, Z Li, J Sun (2015) NaCl-induced changes of ion homeostasis and nitrogen metabolism in two sweet potato (Ipomoea batatas L) cultivars exhibit different salt tolerance at adventitious root stage. Environ Exp Bot149 (2):1141–1153. https://doi.org/10.1104/pp108129494
Yue Y, Zhang M, Zhang J, Duan L, Li Z (2012) SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J Plant Physiol 169(3):255-261. https://doi.org/10.1016/jjplph201110007
Yusuf M, Hayat S, Alyemeni MN, Fariduddin Q, Ahmad A, (2013) Salicylic acid: physiological roles in plants. In Salicylic acid 15-30 Springer, Dordrecht
Zhao WT, Feng SJ, Li H, Faust F, Kleine T, Li LN, Yang, ZM (2017) Salt stress-induced FERROCHELATASE 1 improves resistance to salt stress by limiting sodium accumulation in Arabidopsis thaliana. Sci Rep 7(1):1–16 https://doi.org/10.1038/s41598-017-13593-9
Zheng Y, Jia A, Ning T, Xu J, Li Z, Jiang G (2008) Potassium nitrate application alleviates sodium chloride stress in winter wheat cultivars differing in salt tolerance. J Plant Physiol 165(14):1455-1465. https://doi.org/10.1016/jjplph200801001
Zhou W, He S, Naconsie M, Ma Q, Zeeman SC, Gruissem W, Zhang P (2017) Alpha-glucan, water dikinase 1 affects starch metabolism and storage root growth in cassava (Manihot esculenta Crantz). Sci Rep 7(1):1–17
Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6(5):486–503. https://doi.org/10.1111/j.1467-7652.2008.00336.x
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66(6):675-683. https://doi.org/10.1007/s11103-008-9298-4
Acknowledgments
The authors thank Director, Joint Director (Research), ICAR-Indian Agricultural Research Institute (IARI), New Delhi; Head, Division of Genetics and Incharge, National Phytotron Facility, IARI, New Delhi, for their support provided to accomplish the research activities. Authors also thank Ashwani Kumar Mishra, DNA Xperts Pvt Ltd, Delhi for support in Bioinformatics data analysis.
Funding
This work has been financially supported by Department of Biotechnology, Ministry of Science and Technology (Grant No. BT/PR31301/AGIII/103/1115/2019) and ICAR-Indian Agricultural Research Institute (IARI), New Delhi (Project No. JAN 09 / 16).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DS, MP, and PCS. Performed the experiments: CKS and SS. Analyzed the data: DS, CKS, SS, JK, and DPS. Contributed reagents/materials/analysis tools: MP, PCS, and KG. Wrote the paper: DS, JT, CKS, SS, and KG. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Disclaimer
The funding bodies had no role in the design of the study, data analysis interpretation and writing the manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supp. Fig 1
. Salinity score denoting the effect of salinity stress on tolerant and sensitive cultivars for 5, 7 and 10 d of time interval. Mean values amid same small letters (a, b, c, d) on the bar are non-significantly different by Tukey test at P ≤ 0.05 (JPG 47 kb)
Supp. Fig 2
. Morphological responses of seedling are visualized under (a) control and (b) salt stress condition. (JPG 104 kb)
Supp. Fig 3
. Bar graph representing (a) total chlorophyll content, (b) proline content, (c) soluble sugars, (d) glycine betaine, (e) relative water content, (f) membrane stability index of two cultivars, PDL-1 (tolerant) and L-4076 (sensitive) under salinity stress. Mean values amid same small letters (a, b, c, d) on the bar are non-significantly different by Tukey test at P ≤ 0.05 (JPG 76 kb)
Supp. Fig 4
. Flow chart representing work flow used for generation as well as annotation of contigs from different samples. (JPG 74 kb)
Supp. Fig 5
. De-novo assembly and annotations representing (a) total number of contigs (bp)(b) mapped reads (c) overall read mapping rates for different samples. Biological replicates are named as SL_Treated_1, SL_Treated_1_1, SL_Treated_1_2 for tolerant treated; SL_Treated_2, SL_Treated_2_1, SL_Treated_2_2 for sensitive treated; SL_control_1, SL_Control_1_1, SL_Control_1_2 for tolerant control, SL_control_1, SL_Control_2_1, SL_Control_2_2 for Sensitive control. (JPG 132 kb)
Supp. Fig 6
. Wego plot for showing percentage of genes for different GO terms for tolerant and sensitive cultivar under salt stress condition. (JPG 159 kb)
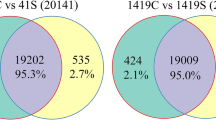
Supp. Fig 7
. Venn diagram representing total number as well as common DEGs in tolerant (1T) and sensitive (2T) cultivars under salt stress. (JPG 37 kb)
Supp. Fig 8
. Total number of SNPs and SSRs detected for different samples. Biological replicates are named as SL_Treated_1, SL_Treated_1_1, SL_Treated_1_2 for tolerant treated; SL_Treated_2, SL_Treated_2_1, SL_Treated_2_2 for sensitive treated; SL_control_1, SL_Control_1_1, SL_Control_1_2 for tolerant control, SL_control_1, SL_Control_2_1, SL_Control_2_2 for Sensitive control. (JPG 129 kb)
Supp. Table 1
Top 20 up regulated DEGs for combination: tolerant treated (T1) and sensitive treated (T2) using EdgeR. (DOCX 17 kb)
Supp. Table 2
. Top 20 down regulated DEGs for combination: tolerant treated (T1) and sensitive treated (T2) using EdgeR. (XLSX 1711 kb)
Supp. Table 3
. Heat map showing differentially expressed genes (DEGs) related to (a) Basic Leucine Zipper Domain (bZIP), Zinc finger motif, WRKY, MYB, basic helix-loop-helix (bHLH, ) transcription factors, phyto-hormones, cell wall modulators, epigenetic modulators andprotein kinases (XLSX 16 kb)
Supp. Table 4
. List of Primers used for validation of NGS data through RT-PCR. (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Singh, D., Singh, C.K., Taunk, J. et al. Transcriptome skimming of lentil (Lens culinaris Medikus) cultivars with contrast reaction to salt stress. Funct Integr Genomics 21, 139–156 (2021). https://doi.org/10.1007/s10142-020-00766-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00766-5