Abstract
A survey and analysis is made of all available ω-gliadin DNA sequences including ω-gliadin genes within a large genomic clone, previously reported gene sequences, and ESTs identified from the large wheat EST collection. A contiguous portion of the Gli-B3 locus is shown to contain two apparently active ω-gliadin genes, two pseudogenes, and four fragments of the 3′ portion of ω-gliadin sequences. Comparison of ω-gliadin sequences allows a phylogenetic picture of their relationships and genomes of origin. Results show three groupings of ω-gliadin active gene sequences assigned to each of the three hexaploid wheat genomes, and a fourth group thus far consisting of pseudogenes assigned to the A-genome. Analysis of ω-gliadin ESTs allows reconstruction of two full-length model sequences encoding the AREL- and ARQL-type proteins from the Gli-A3 and Gli-D3 loci, respectively. There is no DNA evidence of multiple active genes from these two loci. In contrast, ESTs allow identification of at least three to four distinct active genes at the Gli-B3 locus of some cultivars. Additional results include more information on the position of cysteines in some ω-gliadin genes and discussion of problems in studying the ω-gliadin gene family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The wheat seed protein fraction is composed mainly of prolamines—polypeptides rich in prolamine and glutamine, and characterized by solubility in alcohol–water solutions (Shewry et al. 2003). Among the five classes of wheat prolamines, the ω-gliadins are generally considered part of the monomeric gliadin fraction of the wheat seed proteins along with the α- and γ-gliadins. The second major fraction is glutenin—polymeric protein composed mainly of high molecular weight and low molecular weight glutenin subunits (HMW-GS and LMW-GS). This physical–chemical classification is not necessarily an indicator of the evolutionary origin of the five different prolamine classes. The ω-gliadins are likely evolved from a common ancestor to the other gliadins and LMW-GS since the ω-gliadins have similar repeat motifs and are physically tightly linked to the γ-gliadin and LMW-GS genes (Payne 1987; Gao et al. 2007). In contrast, the HMW-GSs are more distantly related to the other wheat prolamines and likely arose from an adjacent globulin-like gene (Anderson et al. 2003). The α- and γ-gliadins and LMW-GS contain cysteines whose disulfide bonds stabilize the polypeptides’ three-dimensional structures while the ω-gliadins are composed almost entirely of a repetitive domain bracketed by short unique sequences generally lacking cysteines. The lack of cysteines would prevent polypeptides from participating in the gluten polymer (Wieser 2007). In addition, the ω-gliadins are thought to also lack methionine resides—thus the reference to the ω-gliadins as being sulfur poor (Tatham and Shewry 1995).
There have been correlations of ω-gliadins to wheat quality (Wang et al. 2008), but the connection is not clear. Altenbach and Kothari (2007) noted that the ω-gliadins respond to variations in nitrogen availability, a factor that could influence the quality of wheats of different genetic basis grown under different environmental conditions. The ω-gliadins have also been implicated in wheat-related allergies (Battais et al. 2005) and an IgE-binding epitope in B-genome ω-gliadins is associated with wheat-dependent exercise-induced anaphylaxis (Matsuo et al. 2005).
Although the ω-gliadins are considered mainly monomeric proteins, fractions of the gluten polymer have been shown to contain subunits with N-termini characteristics of the ω-gliadins. Such subunits have been labeled the D-type LMW-GS (Masci et al. 1993) as part of the gluten polymer fraction along with the A-type glutenin (HMW-GS), B-type glutenin (traditional LMW-GS), and C-type glutenin (small fraction of the α- and γ-gliadins that participate in the polymer) (D’Ovidio and Masci 2004). These D-type glutenins, ω-gliadins with a free cysteine available for crosslinking, have been noted in several studies including Masci et al. (1999) where a cv Chinese Spring ω-gliadin was determined to contain a single cysteine, although the exact location of the cysteine could not be determined.
N-terminal peptide studies have classified the ω-gliadins as ARE, ARQ, KEL, and SRL types (Kasarda et al. 1983). However, evidence suggests that the KEL type arises from the ARQ type by the action of an asparaginyl endoprotease (Dupont et al. 2004). The genes for the first three types are found on the 1A and 1D chromosomes and the encoded proteins are referred to as the ω-1 and ω-2 gliadins, respectively. The SRL-type genes are found on the 1B chromosomes and the encoded proteins are referred to as the ω-5 gliadins (Dupont et al. 2004). Additional post-translational processing is problematic for proteomic determination of the number of distinct genes contributing to the ω-gliadin seed component since a single gene could encode for multiple polypeptides. Attempts to easily classify ω-gliadins by their N-termini is confounded by examples where distinct N-termini seem to originate from more than one genome (Tatham and Shewry 1995; Hassani et al. 2008). There are also reports of a TRQ type (Tatham and Shewry 1995). The present report will focus on DNA sequences and will refer the ω-gliadin genes and polypeptides by the encoded N-termini after signal peptide removal and without additional post-translational processing, e.g., AREL, ARQL, SRLL, and TRQL.
The chromosome organization of the ω-gliadin genes has not been completely determined. The Gli-3 locus is the commonly referenced ω-gliadin locus (Payne et al. 1988) and is tightly linked to the Glu-3 locus (LMW-GS) and near the Gli-1 locus reported to be a compound locus of different gliadin classes, including γ- and ω-gliadins (Payne 1987). A separate ω-gliadin locus (Gli-5) was reported linked to Gli-1 (Pogna et al. 1993), and Metakovsky et al. (1986) reported ω-gliadin genes mapping on both sides of Gli-1.
Only a few ω-gliadin gene sequences have been reported. The sole two reported genomic clones are from either the A or D genome and were restricted to the sequence immediate to the coding regions and thus gave no information on the higher-order structure of any Gli-3 locus (Hsia and Anderson 2001). The other reported ω-gliadin sequences are peptides (Kasarda et al. 1983; Dupont et al. 2000, 2004), PCR products (Masoudi-Nejad et al. 2002; Matsuo et al. 2005; Hassani et al. 2008), partial mRNA sequences and PCR products reported to Genbank but not otherwise published (AY591334, AJ937839, DQ307378, EF116277, EF116278), or assemblies from ESTs (Anderson et al. 2004; Altenbach and Kothari 2007). We previously reported on the general DNA organization of an extended portion of an ω-gliadin gene-rich region from the tetraploid cultivar Langdon (Gao et al. 2007). In that report, we showed that a LMW-GS gene, presumably part of the Glu-B3 locus, lies immediately adjacent to at least a section of the compound Gli-B3 locus.
In the current study, an analysis is made of all available ω-gliadin sequence information and reports on details of the ω-gliadin gene coding regions within the portion of a wheat Gli-3B locus, compares known B-genome ω-gliadin sequences with the different A- and D-genome sequences, mines ESTs for information on transcriptional activity of ω-gliadin genes, proposes model sequences for the Gli-A3 and Gli-D3 loci, and assesses the evidence of cysteine-containing ω-gliadins in wheat.
Experimental
ω-Gliadin genes
A BAC library of T. turgidum ssp. durum (2n = 4x = 28, AABB) cultivar Langdon (Cenci et al. 2003) was screened with γ-gliadin and LMW-GS probes as described previously (Gao et al. 2007). Two BACs, 790O10 and 419P13, contain separate hybridizing bands on Southern analysis for both the γ-gliadin and LMW-GS probes. However, the γ-gliadin hybridizing fragments for BAC 419P13 gave significantly weaker hybridization than those for BAC 790O10. Our previous work (Hsia and Anderson 2001) had shown that γ-gliadin probes will also detect ω-gliadin DNA with weak but still positive hybridization. The two BACs were sequenced and shown to contain either γ-gliadin and LMW-GS genes (790O10) from the A-genome or LMW-GS and ω-gliadin genes (419P13) from the B-genome (Gao et al. 2007). These two BAC sequences can be found at NCBI under accessions EF426564 (BAC 790O10) and EF426565 (BAC 419P13).
Sequencing of BAC 419P13 was by procedures described in detail in Kong et al. (2004). Briefly, randomly sheared BAC DNA was blunt ended with mung bean exonuclease (BioLab), dephosphorylated with shrimp alkaline phosphatase (USB), single A-tailed with Taq polymerase, and the resulting DNA fractionated to 3–5 kb with agarose gels and the Qiagen Gel Extraction Kit. This DNA was used to generate shotgun libraries using the vector pCR4TOPO and transformed into DH10B electroMAX cells (Invitrogen). Randomly picked clones were sequenced at both insert ends with T3 and T7 primers and BigDye chemistry (Applied Biosystems) with an ABI3730xl sequencer.
Sequence analysis began with contig assembly using both Phrap (http://www.phrap.org) and the Lasergene SeqMan module (http://www.DNAStar.com). Gaps and uncertain sequences were resolved by comparing the assemblies from the two software packages and primer walking. Regions of less coverage or ambiguous reads were rechecked with primers designed to cover those regions.
ω-Gliadin EST analysis
Total EST analyses were carried out using direct querying to NCBI (http://www.ncbi.nlm.nih.gov). For individual germplasm queries, EST entries were downloaded from NCBI and placed within databases at the “wEST” site (http://purpurea.pw.usda.gov/wEST) associated with the GrainGenes database (http://wheat.pw.usda.gov). Within the wEST site and GrainGenes are facilities for downloading ESTs from specific wheat germplasms, BLAST querying of those germplasm-specific ESTs, wheat prolamine control sequences within BLAST databases to allow discrimination of BLAST matches to the different prolamine classes, and connections to data on ESTs that have been bin-mapped in wheat. Empirical tests were used to identify BLAST cut-off expectation values for distinguishing specific prolamine classes. For assignment to the ω-gliadin class, it was necessary to interrogate the databases twice, once with an A/D-genome ω-gliadin sequence and a second time with a B-genome ω-gliadin sequence since the two ω-gliadin types are significantly different from each other. The gliadin classes and LMW-GS are similar enough in sequence that some uncertainty can exist with ESTs of short length and/or poorer quality sequence. There was usually a range of BLAST hits borderline between those ESTs with definitive assignments and ESTs from other prolamine classes. In these cases, 10–30 ESTs at this boundary were manually inspected to confirm assignments. Probes and NCBI accessions for interrogating databases for genomic or EST sequences are as follows: α-gliadin, U08287; γ-gliadin, AF234646; A- and D-genome ω-gliadin, AF280605; B-genome ω-gliadin, AB181300; LMW-GS, U86028.
ESTs were assembled into contigs using known gene sequences, when available, as scaffolds for the assemblies. Assemblies for ESTs from individual wheat cultivars used Clustal V and Clustal W implementations in the Megalign and Seqman software modules of the Lasergene II package (DNAstar, Inc.) followed by visual inspection of alignments and manual trimming of terminal vector sequences and sequence adjustments when necessary. Chimeric sequences were identified by individually analyzing ESTs which joined alignments but included a sequence region that totally diverged from the rest of the alignment or those ESTs that were identified by BLAST as containing target sequences but did not join contigs. The non-ω-gliadin portions of such chimeric sequences were discarded and those ω-gliadin portions longer than 200 bp were used in EST assemblies. Assemblies for consensus full-length DNA and amino acid sequences using ESTs from all germplasms were carried out manually since software was unable to handle the combinations of highly repetitive DNAs, irregular deletions of repetitive regions, and allelic differences in sequences. In these cases, a consensus sequence required a minimum of three independent EST sequences across all regions of the consensus.
ESTs produced by Dupont Corporation are believed to have originated from a spring wheat line of uncertain identity and will be referred to as the “Dupont” set instead of by cultivar.
Chromosome assignments
To map ω-gliadins to specific wheat chromosomes, PCR primers were designed based on single nucleotide polymorphisms (SNPs) among grouped sequences. Specific primers were designed so that the 3′ terminal nucleotide corresponds to the site of the SNP, and an artificial base pair mismatch was introduced next to the 3′ end, as described by Hayashi et al. (2004). Such primers were used to map ω-gliadins to specific chromosomes by amplifying DNA from the Chinese Spring wheat nulli-tetrasomic aneuploid lines of homoeologous group 1, i.e., nullisomic 1A-tetrasomic 1B (N1AT1B), nullisomic 1B-tetrasomic 1D (N1BT1D), and nullisomic 1D-tetrasomic 1A (N1DT1A). PCR amplification was conducted in a 20-µl volume containing 50 ng genomic DNA template, 0.2 µM of each primer, 0.25 mM dNTPs, 0.5% DMSO, 1× PCR buffer (Promega), and 0.5 U Go Taq DNA polymerase (Promega). The amplification program was 94°C for 5 min for initial denaturation followed by 35 cycles of 94°C for 30 s, annealing for 30 s, and 72°C for 1.5 min.
Primers were designed from clusters of related ω-gliadin sequences as described in ‘Results and discussion’. Forward primer EF116277-f2 (GCAACAACCATTCCCCCAACACT) is specific for ω-gliadins EF116277, AF280606 (gene G3), and an ARQL-type consensus sequence. Forward primer AY667097-f2 (AGATCGCCACTGCCGCTAGTG) is specific for AY667097, AF280605 (gene F20b), and an AREL-type consensus sequence. The reverse primer AY667097-r1 (CATTGGCCACCGATGCTTGTAAG) is derived from the conserved 3′ end of ω-gliadins and can be used for all of the aligned genes. Primers AA-f2 (ACAATCATATCCGCTGCCA) and AA-r2 (ATGGTCATTGTGGTTGTAGGAG) are specific for AB059812, AY591334, and DQ307378.
Results and discussion
B-genome ω-gliadin genes
We have previously reported the general organization of a genomic 139,403-bp BAC clone containing multiple B-genome ω-gliadin sequences (Gao et al. 2007). We now report on the detailed structure of those B-genome ω-gliadin sequences. Figure 1 shows the positions of eight ω-gliadin sequences in BAC clone B419P13 (Genbank accession EF426564). B-genome ω-gliadins are commonly referred to as the ω-5 group of ω-gliadins (Dupont et al. 2000; Matsuo et al. 2005). For consistency, the ω-gliadin sequences of Fig. 1 will be assigned the following nomenclatures, i.e., ω-5-F1, ω-5-ψ1, ω-5-1, etc. To be more readable in the current report, further references to these sequences here will omit the ω-5, i.e., “1” will refer to full-length gene “ω-5-1.” In the future, nomenclatures of further prolamine genes should include the germplasm of origin, i.e., ω-5-1(LDN), where LDN is the formal abbreviation of the wheat tetraploid cultivar Langdon (GrainGenes database; http://wheat.pw.usda.gov). Researchers needing new Triticeae germplasm abbreviations should apply to GrainGenes at curator@greengene.cit.cornell.edu.
Organization of part of a Gli-B1 locus. A section of a Gli-B1 locus containing eight ω-gliadin genes and gene fragments was cloned in BAC 419P13 (EF426565). The relative positions of the ω-gliadin sequences are shown and includes two complete ORFs (1, 2), two pseudogenes (Ψ1, Ψ2) and four 3′ ω-gliadin fragments (F1–F4). The DNA is shown in reverse orientation from the Genbank entry, and the ω-gliadin positions in EF426565 at stop codons for full-length genes and approximate positions for fragments are as follows: F1, 106,600; Ψ1, 76,749; Ψ2, 66,558; F2, 47,500; 1, 41,831; F3, 21,900; 2, 16,212; F4, 7,800. Arrows under ω-gliadin sequences indicates potential directions of transcription. The arrowhead at the left end of the DNA fragment indicates extensive DNA sequence is known and does not include more ω-gliadin sequences. The multiple arrowheads on the right indicates the direction of non-sequenced DNA that may contain more ω-gliadin sequences
Of the eight ω-gliadin sequences in Fig. 1, only two (1 and 2) are assumed to be active ω-gliadin genes since they contain similar uninterrupted ORFs and flanking DNAs consistent with active genes. The mature ω-gliadin protein encoded by gene 1 would have 432 amino acid residues and a molecular weight of 51,360 Da, and gene 2 would encode an ω-gliadin of 419 amino acid residues and 50,887 Da. These sizes are consistent with the B-genome ω-gliadin sizes of 48,900–51,500 determined by mass spectroscopy by Dupont et al. (2000). Genes 1 and 2 also contain the mature polypeptide start SRLL characteristic of B-genome ω-gliadins.
Sequences ψ1 and ψ2 contain flanking DNAs consistent with active genes, but their coding regions include seven and two premature stop codons, respectively. In addition, gene ψ1 contains four frameshifts. Without stop codons and frameshifts, ψ1 would encode a polypeptide of about 467 amino acid residues that would have a molecular weight of about 56,784 Da. Gene ψ2 is missing most of the typical ω-gliadin repetitive domain and, if it were an active gene, would encode a polypeptide of about 110 amino acids and 13,092 Da—smaller than any reported ω-gliadin and potentially missed in protein analyses.
In addition to the four ω-gliadin genes (by the criteria of start and stop codons and appropriate immediate flanking DNAs), there are also four ω-gliadin gene fragments scattered throughout this genome region (Fig. 1; F1–F4). All four contain the final section of a repetitive domain plus varying lengths of 3′ non-coding sequence. No non-ω-gliadin genes are detected interspersed with the eight ω-gliadin sequences.
The length and similarity of different ω-gliadin DNA sequences is shown in the dot plots of Fig. 2 comparing gene 1 to other ω-gliadin DNA sequences. Gene 1 is very similar to AB181300 of Matsuo et al. (2005)—the solid block-like structure in Fig. 2a is the result of comparing the closely related repeat motifs across the repetitive domain. A similar but smaller block is evident in comparison to gene ψ2 (Fig. 2b). Note also that the sequence similarities extend both 5′ and 3′ to the start and stop codons (arrows in Fig. 2). In contrast, only the stop codon is present in the four truncated ω-gliadin gene fragments (example in Fig. 2c).
Comparing ω-gliadin sequences. Gene 1 from Fig. 1 is compared by dot plots to other ω-gliadin sequences to include coding and near flanking regions. a 1B ω-gliadin (AB181300) of Matsuo et al. (2005). b pseudogene Ψ2 of Fig. 1. c ω-Gliadin fragment F1 of Fig. 1. d ω-Gliadin F20b (AF280606) of Hsia and Anderson (2001). Matching criteria were 80% over a 20-base window. Down arrows indicate start codons and upward arrows indicate stop codons
All eight ω-gliadin sequences are in the same transcription orientation (arrows in Fig. 1) and analysis of the entire sequence of BAC B419P13 indicates multiple rounds of duplication lead to this set of ω-gliadin genes and gene fragments (Gao et al. 2007). Whether these sequences represent the entire B-genome ω-gliadin locus is not determined. As indicated in Fig. 1, no additional ω-gliadin sequences exist immediately left in the figure. However, no further sequence is known past the right end of the BAC and additional ω-gliadin gene members of the Glu-B3 locus may exist since analysis of 1B ω-gliadin ESTs indicates at least three to four active genes in some cultivars (see below).
In addition to the four genes reported here (two active and two pseudogenes), only two other B-genome ω-gliadin gene sequences are found in Genbank, i.e., the two PCR fragments (AB181300 and AB181301) reported by Matsuo et al. (2005) with BLASTN matches to gene 1 of e = 0. An additional match is to cDNA M11336 at e value of 2e −86. Although M11336 is listed at NCBI as a γ-gliadin, on close examination, M11336 is found to be a chimeric clone starting with two overlapping short fragments of the 3′ end of B-genome ω-gliadin sequences fused to a partial LMW-GS sequence (not shown). The only other match is to Genbank entry DQ317535—a cv Langdon PCR-generated sequence only one base different from the current gene 2 and likely an PCR amplification from that gene.
The encoded proteins for all known full-length B-genome ω-gliadins are shown in the amino acid alignment in Fig. 3. Genes 1 and 2 are similar but not identical to the two sequences reported by Matsuo et al. (2005) from cultivar Norin 61. Although tetraploid Langdon and hexaploid Norin 61 wheats both have two ω-gliadin sequences most similar to each other, these two pairs are not direct orthologous pairs. The two Langdon sequences share an additional repeat unit (FPQQQ at position 101) with respect to the two Norin 61 sequences. In addition, the two Langdon sequences share differences to the two Norin 61 sequences at four positions, i.e., the Langdon sequences are missing a glutamic acid (E) codon found in both Norin 61 sequences at position 35, and differences in amino acids exist at positions 128, 376, and 435. Three similar synonymous DNA base changes also distinguish the two pairs (not shown). Thus, the two Norin 61 and Langdon ω-gliadin gene pairs likely arose as separate duplication events from an orthologous gene in two distinct ancestor germplasms. Additional explanations not ruled out are gene conversions or that the directly orthologous genes exist but have not yet been identified.
Full-length 1B ω-gliadin amino acid sequences. All known full-length 1B ω-gliadin DNA coding sequences are converted to amino acid sequences and aligned. Amino acid differences are shaded. Sequences are defined to be full length if they have an ORF and similar non-coding flanking DNAs, and include the two active (1, 2) and two pseudogenes (Ψ1, Ψ2) of Fig. 1 (EF426564), and includes one active (AB181300) and one pseudogene (AB181301) of Matsuo et al. (2005). The vertical bar indicates the junction of the signal peptide and the mature ω-gliadin sequences. Arrowheads below the Ψ1 sequence indicate frameshifts in the original DNA sequence. Sequences are named by Genbank accession and numbering from Fig. 1
Both genes Ψ1 and Ψ2 are thus far unique and there is no evidence of closely related active genes as evidenced by matching ESTs. In addition to individual base changes, gene Ψ1 has an additional 63 amino acid section in the repeat domain. This region is very similar, though not identical, to the immediate N-terminal region of the other three longer B-genome ω-gliadins—suggesting a 189-bp tandem duplication that has undergone some sequence divergence since the duplication. Gene Ψ2 is the shortest known ω-gliadin sequence and probably resulted from a large deletion in the repetitive domain. It is not known if gene Ψ2 was in its current state, with two premature stop codons, before such a deletion. However, the large ω-gliadin protein reported by Gianibelli et al. (2002) and gene Ψ2 suggest caution in looking to identify ω-gliadin proteins only in the 35–55-kDa size range where ω-gliadins have been previously reported.
Comparisons to A- and D-genome ω-gliadin genes
The dot plot in Fig. 2d compares B-genome gene 1 to an ω-gliadin only previously known to be from either the A- or D-genome (Hsia and Anderson 2001). This comparison shows less similarity than to the better match to another B-genome ω-gliadin gene (Fig. 2a). A major distinguishing feature of the B-genome ω-gliadins is a different repeat motif pattern compared to the A- and D-genome ω-gliadins, i.e., the B-genome pattern is mainly based on FPQ2–4 (in nearly half the repeats the F is replaced by I, L, or occasionally P or S) and the A- and D-genome pattern is based on PFPQ1–2PQQ. The different patterns are seen in Supplementary Fig. 1S, and the differences between genes 1 and Ψ1 (B-genome) and genes F20b and G3 (A- and D-genomes) are evident. One result of the different repeat base motifs is that there are more, but shorter, repeat units in the B-genome genes. This difference in repeat patterns is assumed the result of drift in the repetitive domain after the separation of the B-genome ancestor from a common ancestor of the A- and D-genomes.
The A- and D-genome ω-gliadin protein repeat pattern is also similar to the repeat pattern of the wheat γ-gliadins (PFPQ1–2(PQQ)1–2) and is the basis of the stronger cross-hybridization of γ-gliadin and A/D-genome ω-gliadin DNA sequences than other prolamine class pairs—including the B-genome ω-gliadin sequences. At the amino acid level, the differences are even more pronounced. In the pairwise dot-plot comparisons of the protein sequences of an A/D-genome ω-gliadin, a B-genome ω-gliadin, and a γ-gliadin, the γ-gliadin sequence is more similar to both ω-gliadins than the two ω-gliadin types are to each other (Supplementary Fig. 2S).
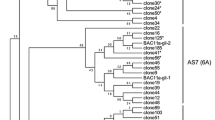
The DNA coding regions for all available A- and D-genome ω-gliadin genes are aligned (Supplementary Fig. 3S). No derived amino acid alignments were attempted since many of these genes have multiple frameshifts. However, there is a clear separation of the sequences into two classes. This separation is seen in Fig. 4 where all available ω-gliadin coding sequences are used to construct a phylogenetic tree. The resultant tree is a composite of separate alignments of the B-genome and A- plus D-genome ω-gliadin sequences since the repeat structure of 1B ω-gliadins is too diverged for realistic alignments with 1A and 1D ω-gliadin sequences. It is assumed that these two branches had a common ancestor—indicated by the left-most node. Within the A/D branch is a fork leading to branch A containing a cluster of four sequences including one that has been assigned to the A-genome (AB059812-Masoudi-Ψ; Masoudi-Nejad et al. 2002). This A-genome assignment has been confirmed using primers unique to branch A and amplifying DNA from Chinese Spring nulli-tetrasomic wheat lines (Fig. 5a). All four of these A-genome ω-gliadins are pseudogenes, i.e., contain premature stop codons and one or more frameshifts. While there may be active genes in this family, neither any matching EST sequences nor an ARHL start of a mature ω-gliadin protein have been reported.
Phylogenetic tree of ω-gliadin DNA sequences. Reported ω-gliadin DNA sequences are aligned separately for the 1B and 1A/D sequences by Clustal V and the resulting two branches are shown connected by theoretical relationships to a common ancestral ω-gliadin sequence (left-most node). Names are by Genbank accession, gene name (if named), first author of report (if published) and indication of being a pseudogene (Ψ). The four B-genome gene sequences and two consensus A/D-genome gene sequences from the current report are all shown in bold. Genomes or origin are indicated by A, B, D, or A/D (1A and/or 1D). AJ937839 is indicated as a pseudogene by sequence, but may include sequence artifacts of cloning
Genome assignments of ω-gliadin sequences to A, B, and D genomes. Clusters of ω-gliadin sequences as indicated in Fig. 4 were analyzed for unique DNA priming sites. Specific PCR primers were designed and used to amplify DNA from genomic DNA of wheat cv Chinese Spring nulli-tetrasomic genetic stocks. Primers are shown in Supplementary Fig. 3Sa. a Primers for branch A. b Primers for branch D′. c Primers for branch A′. Primer names are below each frame. Predicted size of amplified fragments from DNAs within each branch were used to design primers are shown below each frame. The DNA ladder was the 100-bp DNA Ladder from Invitrogen (Cat. No. 15628-019)
The second fork from the A/D branch in Fig. 4 leads to two suggested groupings of sequences containing all other reported A/D ω-gliadin DNA sequences—plus two labeled the AREL- and ARQL-consensus sequences derived from ESTs as described and discussed in detail below.
Complication in studying the wheat ω-gliadin genes
The wheat ω-gliadin genes are an elusive subject for detailed research, and anyone studying these genes needs to be aware of major complicating factors—among which are technical difficulties in cloning these highly repetitive DNA sequences, chimeric ESTs, the high frequency of pseudogenes, and the difficulty in estimating both the total and active numbers of ω-gliadin genes.
The difficulty in cloning ω-gliadin genes has been reported previously, i.e., where recombination-deficient host bacteria and a vector that is less-susceptible to insert deletions were necessary to clone two genomic fragments each containing an ω-gliadin gene (Hsia and Anderson 2001). In that study, initial attempts to generate subclones of the original fragments resulted in numerous deletions of various sizes, even when the clones were grown in recombination-deficient hosts. Similar problems in obtaining stable ω-gliadin clones and subclones have also been noted elsewhere, e.g., Hassani et al. (2008). This tendency for clones to delete portions of the ω-gliadin repetitive domain requires caution with either cloning or PCR amplification of sequences from this gene family as well as all the prolamines. Although the root cause of this tendency is not confirmed, it is assumed that it is related to the nature of the ω-gliadin repetitive domain. Almost the entire coding sequence is composed of a series of short DNA repeats encoding either PFPQ1-2PQQ (A- and D-genomes) or FPQ2–4 (B-genome). Within the ω-gliadin DNA repetitive sequence is found numerous occurrences of the sequence GCTGGGGG on the anti-sense DNA strand—a one-base-change difference from the strand-pair Chi recombination recognition sequence GCTGGTGG that is recognized by the RecBCD enzyme integral to bacterial homologous recombination (Bianco and Kowalczykowski 1997). The CCCCCAGC complementary coding strand sequence is found seven times in ω-gliadin sequence F20b (AF280605), ten times in ω-gliadin gene 1 of Fig. 1 (EF426564), but not at all in examples of α-gliadins (X01130), γ-gliadins (AF234646), LMW-GS (U86030), or HMW-glutenins (X12928).
Whatever the basis of ω-gliadin sequence instability, their study should be approached with caution. It is especially important to minimize replication of the sequences and maintain clones in hosts as recombination deficient as possible. PCR should be carried out with special care since so many replications occur even before cloning. In addition, we suggest other minimal guidelines involving PCR for all prolamine studies, especially for the ω-gliadins, i.e., initial PCR products must match the size and sequence length of cloned PCR reactions, sequencing of multiple PCR clones, and different clones must come from different PCR reactions.
Once ω-gliadin sequences have been cloned and sequenced, a large number of inactive genes have been reported. Figure 4 shows that of the 15 reported ω-gliadin gene sequences, nine are pseudogenes, i.e., genes that are identifiable but assumed to be inactive since their sequences contain combinations of functional lesions such as premature stop codons and frameshifts. Even this number of inactive genes could be conservative. Two of the “active” genes are derived from PCR reactions of coding sequence only (AB181300, AY667097) and therefore do not assure intact promoter and other controlling elements. Neither is the apparent presence of significant conserved flanking DNA in three sequences (AF280605, genes 1 and 2 of EF426564) definitive proof that the genes are actually leading to ω-gliadin proteins. Only the incomplete coding region cDNA clone AF937839 is assured of originating from an active gene, and this case is somewhat anomalous or artifactual since the original sequence may be a chimera between ω-gliadin types, has a short non-matching 5′ end, and has a frameshift near the 3′ end.
Pseudogenes are known to be common in cereal prolamine gene families, e.g., wheat α-gliadins (Anderson 1991), the first ω-gliadin sequence report (Hsia and Anderson 2001), and maize where a 22-gene zein cluster contains seven active and 15 pseudogenes (Song et al. 2001). The most common defect in prolamine pseudogenes are premature stop codons, and most of these occurring at original glutamine codons. Since glutamine is encoded by the two codons CAA and CAG, the common transition C→T can create TAA and TAG stop codons. With their large repetitive domain and high frequency of glutamine codons, the ω-gliadin genes are likely even more susceptible to premature stop codon generation than other prolamines. Including all codons, 40% of the codons of gene F20b (AF280605) are a single base change from becoming a stop codon. For the two intact B-genome ω-gliadin genes reported here, 53% of the codons might become stop codons with a single base change. In contrast, triticin is a large, non-prolamine wheat storage protein whose gene (EU482414) is less at risk with only 20% of its codons potential stop codons with a single base change. For larger ω-gliadin variants, such as reported by Gianibelli et al. (2002), the risk rises to about 70% of the codons. Thus, the ω-gliadin genes will particularly tend to inactivate the larger they become.
There is also a curiosity in the codon distribution in ω-gliadin genes from all three hexaploid wheat genomes. For the other major wheat prolamine classes (HMW- and LMW-GS, α- and γ-gliadins), the CAA/CAG ratio of glutamine codons is around 2:1, but for the ω-gliadins it is 7:1. Why CAA is so much more prevalent in the ω-gliadins than among the other wheat prolamines is unknown. One consequence of higher frequencies of one codon in homopolymer runs is increased opportunity for unequal recombinations and/or slip-mismatching during replication. These would result in block changes ranging from single repeat unit to large section deletion/duplications of the repetitive domain.
The difference in the repetitive structure in B- versus A/D ω-gliadins is also a technical complication in studying this gene family. For example, if a single probe is used in Southern hybridization of restricted genomic DNA, then the two distinctive repetitive domain patterns (A/D vs. B) will cause under-detection of one pattern, i.e., a D-genome ω-gliadin probe will return stronger hybridization signals from the A- and D-genome genes than from the B-genome genes, and vice versa. A database resource at http://wheat.pw.usda.gov/wEST permits BLAST analysis of EST collections from specific wheat and barley cultivars. Included with the EST databases as a control are known prolamine sequences to aid assessing BLAST expectation values and score cut-offs among the different prolamine classes. A BLASTN search of 352,133 wheat cv Chinese Spring sequences found that the Langdon gene 1 B-genome ω-gliadin sequence matches to A/D ω-gliadin sequences at only an expectation value of e −16 compared to an e = 0 match to the 1B gene of Matsuo et al. (2005). In comparison, an α-gliadin gene sequence (X01130) matches to α-gliadin sequences starting at e = 0, LMW-GS sequences at e −11, and γ-gliadin and ω-gliadin sequences at e −6. Since we have shown that the gliadin/LMW-GS sequences will not detectably cross-hybridize under standard Southern analysis conditions (Anderson et al. 1997), using a B-genome ω-gliadin probe will not significantly detect A/D-genome genes via hybridization, and vice versa. Similarly, isolation of ω-gliadin sequences from genomic or cDNA libraries may need two different probes to identify genes of all three hexaploid genomes. These results also indicate that, at least for sequence similarity analyses, the different motifs of the A/D ω-gliadin sequences are distinct enough from the B-genome ω-gliadin sequence motifs that the A/D- and B-genome ω-gliadins are almost different prolamine classes and related more by their evolutionary history than their sequences would indicate.
Pseudogenes and gene fragments further complicate estimating copy numbers of active genes by Southern analysis of genomic DNA. Sabelli and Shewry (1991) estimated 15–18 copies of ω-gliadin type sequences in cv Chinese Spring and five to ten copies in the tetraploid cv Langdon. If similar organizations exist at the Gli-A3 and Gli-D3 loci as for the Gli-B3 locus region reported here, then the pseudogenes and gene fragments would cause the number of active genes to be only a fraction of the estimated total sequence numbers.
ω-Gliadin A/D genome ESTs
ESTs are single sequence reads from cDNA clones and individually are of variable sequence quality. However, when sufficient EST resources are available, multiple reads across the same region will allow consensus calling for reliable DNA sequences. The large wheat EST resource in Genbank was mined for ω-gliadin sequences. One complication that became apparent is the high frequency of problem EST entries containing ω-gliadin sequences. Careful individual analysis showed that about half of the ESTs containing ω-gliadin sequences were aberrant. Most were chimeras of an ω-gliadin sequence and one or more other distinct genes (analyses not shown). Particularly common were chimeras of ω-gliadin fused to γ-gliadin sequences with the fusion point within their repetitive domains—possibly due to a combination of the similarity between the repetitive domain motifs in these two prolamine classes and the presence of numerous three to four tandem CAA codons. In addition, apparent deletions of sections of repetitive domains were found in a minority of ω-gliadin ESTs. An EST was judged chimeric or contained a cloning-derived deletion if its sequence did not match any other sequence within Genbank, i.e., each chimera or deletion was judged the result of a unique event during the cloning process. Approximately 50 such ESTs were checked, and all were unique. This does not establish that none were authentic, but does support that no specific such sequence is a major constituent of the wheat endosperm mRNA complement or is a major factor in wheat quality.
Wheat ω-gliadin sequence-containing ESTs were individually analyzed for authentic ω-gliadin sequences. Those ESTs that had less than 200 bp of ω-gliadin sequence or had multiple frameshifts were discarded. Chimeric ESTs with long ω-gliadin intact reading frames were used when the ω-gliadin segment was clearly distinguished from other sequences that were removed. No individual cultivar had ESTs spanning a complete coding region. However, combining 1A and 1D EST DNAs of different cultivars generated two proposed complete coding sequences (ESTs are listed in Supplementary Table 1S). These ω-gliadin ESTs were converted to amino acid sequences and assembled into two protein patterns as shown in Fig. 6. One composite mature protein begins with the AREL N-terminus (Fig. 6a) and the second with ARQL (Fig. 6b). Note that these two consensus amino acid sequences are composites from several cultivars and models for two classes of ω-gliadins and not actual gene sequences.
AREL and ARQL consensus amino acid sequences. Derived amino acid sequences from the ESTs listed in Supplementary Table 1S were assembled into full-length ω-gliadin protein sequences. a The underlined sequence in the AREL consensus marks the sequence not present in Cheyenne gene F20b and the boxed region matches Cheyenne EST BE422668. b The two boxed regions of the ARQL consensus match the peptide sequences of Masci et al. (1999). Unique cysteine and methionine residues are circled
The accuracy of the two consensus sequences was checked by comparing to the limited available known sequences. Hsia and Anderson (2001) reported an apparently full-length AREL-type ω-gliadin gene (F20b) from cv Cheyenne. The encoded F20b protein would match the AREL consensus sequence except for a 113 amino acid block present only in the AREL consensus (underlined sequence). There is also a single cv Cheyenne EST the spans most of this region (BE422668) and that matches the consensus (boxed in Fig. 6a), including most of the apparent deletion region of F20b. EST BE422668 matches the F20b DNA sequence exactly over the 288 bp of their overlap, and the EST matches exactly to the 573 bp overlap with the AREL consensus DNA (consensus DNA sequences are given in Supplementary Fig. 4S). The latter further confirms the validity of the consensus sequences.
For the ARQL consensus, there is no full-length gene sequence for comparison. However, Masci et al. (1999) reported that cultivar Chinese Spring contained a gene for an ω-gliadin with peptides that match the ARQL consensus (blocked sequences in Fig. 6b). These two peptide segments match the encoded protein of the ARQL consensus as indicated, and do not match the AREL consensus protein. In addition, although the ω-gliadins have been considered as lacking cysteine or methionine residues, the ARQL consensus also confirms the peptide finding of Masci et al. (1999) of both single cysteine and methionine residues in Chinese Spring ESTs (circled in Fig. 6b). This cysteine residue is found in some, but not all germplasms (see below). The ARQL consensus amino acid sequence also matches, with two residue exceptions, a tentative consensus sequence (TC262770) from The Institute for Genomic Research reported by Altenbach and Kothari (2007). However, the exact derivation of TC262770 is no longer available.
ESTs from individual cultivars were examined for encoded ω-gliadin sequences. Most ESTs were from the 3′ region. Those ESTs with 5′ encoding sequences were usually unique chimeric clones. Multiple ESTs encoding polypeptides matching the AREL and ARQL models were identified from cultivars Chinese Spring, Cheyenne, Butte 86, Glenlea, and Recital (Supplementary Table 1S). For Chinese Spring, 16 and 14 ESTs encoded polypeptides matching the AREL and ARQL types, respectively. For Recital, there were 22 matches to the AREL type, but only two to the ARQL type. For the other three cultivars, there were three to nine ESTs divided between the two types. Sequences from different cultivars match either of these two consensuses, and in no case is there yet evidence for multiple sequences from a cultivar for either of these patterns, i.e., two unique sequences per cultivar, one each for the AREL and ARQL patterns, although there were some cultivar-specific differences such as single amino acid or repeat unit differences (not shown).
A TRQL ω-gliadin type has been reported by Tatham and Shewry (1995) and Gianibelli et al. (2002) originating from T. monococcum and T. tauschii, respectively. The only known DNA sequences that would encode this motif are gene Ψ1 of the current report and four short ESTs of poor sequence quality that are part of the Dupont EST set. The pseudogene would not be expected to produce full-length ω-gliadins, and the four ESTs do not exactly match the N-terminal sequences of Tatham and Shewry (1995) and Gianibelli et al. (2002). Thus, confirmation of TRQL-type ω-gliadins is still lacking.
A- and D-genome assignments
The ARQL-type ω-gliadins have been assigned to the A-genome by Dupont et al. (2004) and Kasarda et al. (1983). AREL ω-gliadins have been reported to be D-genome in several studies (Kasarda et al. 1983; Dupont et al. 2000; Hassani et al. 2008). The only confounding reports are Masci et al. (1999) and Tatham and Shewry (1995) where an ARQL-type sequence is reported from the D-genome and Ae. speltoides (B-genome ancestor), respectively. To test genome assignments, DNA sequences were used to design specific primers that should only amplify sequences within branches A, A′, and D′ of Fig. 4. The results confirm that the Masoudi-Nejad et al. (2002) sequence is from the A-genome (Fig. 5a), and shows that the three other genes in branch A of Fig. 4 are also from the A-genome. Only a single band was amplified, and at the expected size—which indicates the A branch may be represented by a single gene and the members of branch A are alleles of this gene. The ARHL sequence reported by Masoudi-Nejad et al. (2002) is a member of the branch in Fig. 4 thus far composed entirely of inactive genes. In addition, no ESTs exist in Genbank that have either the ARHL motif or match the signal peptide sequences common to the sequence of branch A. Until contrary evidence is found, it is assumed these PCR products are from an inactive branch of the ω-gliadin gene family and make no contribution to the wheat seed protein complement.
The next objective was to determine if genome assignments could be made for the two clusters of sequences in the A′/D′ branch of Fig. 4. Specific primers were designed for each of the two clusters and the results are shown in Figs. 5b and c. Primers EF116277f2 and AY667097r1 amplified two bands as shown in Fig. 5b. These are of sizes appropriate to the original sequences used for primer design and the faster and brighter fluorescent band may represent two fragments of nearly the same size. Since no fragments amplify from the A-genome, this cluster of ARQL-type sequences is assigned to the A-genome that contains at least two to three active and inactive genes.
Similarly, the branch containing the AREL consensus sequence was analyzed using specific PCR primers. As shown in Fig. 5c, no fragment was amplified from the N1DT1A line, but single band results from amplifying from lines that contain the 1D chromosome. Thus, the branch containing the AREL consensus and three previously reported sequences are assigned to the D-genome. Since only a single band was amplified, either there is only a single matching AREL ω-gliadin gene in cv Chinese Spring or multiple genes have almost identical sequences. All primer sequences are given in Supplementary Fig. 4aS, and priming sites in the three classes of A/D-genome genes are shown in Supplementary Fig. 4bS.
One unanswered question is the number of distinct ω-gliadin proteins in the wheat endosperm. The number of different ω-gliadins in the wheat endosperm cannot be determined based on existing data. Dupont et al. (2006), in a study of the cultivar Butte 86, report six ω-gliadins, i.e., two for the A-genome, three for the B-genome, and one for the D-genome—results that agree with the current report. Dupont et al. (2004) also found at least three ω-gliadins from a T. urartu A-genome and two from the Chinese Spring 1A chromosome. Seilmeier et al. (2001) report six to nine ω-gliadins, depending on individual germplasms analyzed. The apparent single amino acid sequence pattern for both AREL and ARQL types within single cultivars does not necessarily conflict with reports of multiple A- and D-genome ω-gliadins. The EST AREL and ARQL consensus amino acid sequences suggest a limited number of ω-gliadin patterns, but the limited number of quality EST sequences from individual cultivars does not rule out multiple genes of very similar sequence. Even if AREL and ARQL consensus sequences indicate single genes, differential post-translational N-terminal processing of ω-gliadins could lead to two or more distinct polypeptides per gene.
ω-Gliadin B-genome ESTs
Only cultivars Chinese Spring and Recital have enough ESTs for reasonable 1B ω-gliadin sequence assemblies. Cultivars Butte 86, Cheyenne, Glenlea, and Wyuna have too few ESTs to assemble reliable consensus sequences, but all seem to have at least two species of 1B ω-gliadins (not shown). The Chinese Spring and Recital 1B ESTs used to form assemblies are listed in Supplementary Table 1S, and the derived amino acid sequences are shown in Fig. 7. Since only 3′ ESTs are abundant enough to form assemblies, only those 3′ regions are compared to the same regions in the three only known full-length 1B ω-gliadin protein sequences. Three unique Chinese Spring 1B ω-gliadins are found (Chinese Spring 1–3 in Fig. 7), with 16, 27, and 19 ESTs, respectively. The three sequences are very similar—consistent with the similar pairs of 1B sequence from Gao et al. (2007) and Matsuo et al. (2005).
Multiple 1B ω-gliadins in cultivars Chinese Spring and Recital. Derived amino acid sequences from ESTs of cultivars Chinese Spring and Recital were assembled in three and four distinct 1B ω-gliadin sequences, respectively, and compared to the derived amino acid sequences of the only three known intact 1B ω-gliadin sequences. Amino acid residue differences are shaded. Horizontal lines separate gene from EST assembly sequences. ESTs are listed in Supplementary Table 1S. Genes 1 and 2 are as in Fig. 1. The peptide sequence below the C-terminus of Recital 4 is the sequence if the frameshift were corrected
The Recital EST-derived amino acid sequences assemble into four distinct polypeptides (Recital 1–4) supported by five, seven, 15, and 16 ESTs, respectively (Supplementary Table 1S). The sequence of Recital 2 contains a cysteine residue near the C-terminus—supported by 16 ESTs reading in both orientations. The five ESTs encoding the Recital 4 sequence all contain a frameshift near the end of the repetitive domain. This would result in the Recital 4 polypeptide having an abnormal ω-gliadin C-terminus. For comparison, the Recital 4 sequence in Fig. 7 also shows the encoded C-terminus without the frameshift. Since the Recital ESTs originated from normalized cDNA libraries (P. Leroy, personal communication), the Recital alignments were examined for suggestions of duplicate sequences. Although the alignments did not indicate such duplicate reads, further examination of non-normalized Recitals ESTs/mRNAs is suggested.
Cysteine residues in ω-gliadin sequences
As previously described, Masci et al. (1999) identified a Chinese Spring 1D-encoded ω-gliadin with a single cysteine residue. The location of the single cysteine was not determined by Masci et al. (1999), but was proposed to be more terminal than the larger amino acid sequence fragment—consistent with the Chinese Spring cysteine at 31 residues C-terminal to that larger fragment boxed in Fig. 6b. This cysteine is circled in Fig. 6b and is found in five ESTs of cv Chinese Spring (BG263062, BJ235153, BJ232465, BJ291572, CJ635325), three from Butte 86 (BQ804207, BQ804424, BQ804665) as reported by Altenbach and Kothari (2007), and one from Recital (CDS896637). In contrast, a phenylalanine residue is encoded at that position by several ESTs from cvs Cheyenne and Glenlea (not shown). Since a TTT triplet (Phe) is a common component of ω-gliadin repetitive units, it is assumed a single T-to-G transversion event at this position resulted in the TGT (Cys) codon likely occurred once in an ancestral gene. No cysteine-encoding ESTs at this position were found in other cultivars, but the number of ESTs is too few to be definitive. Thus, in the case of cv Recital, there are at least two cysteine-containing ω-gliadins, one each from the Gli-B3 (Recital 2) and Gli-D3 (AREL-type) loci.
The possibility of other positions is indicated in several other sequences, but the number of sequences is too few for confirmation. For example, the truncated pseudogene Ψ2 would include a single cysteine residue if the gene were active, but is at a position thus far unique among known ω-gliadin sequences. In addition, the cv Wyuna EST BQ608902 and PCR fragment EF116278 each would encode unique cysteine residues, but both are only supported by single sequences and have sequence defects that question their reliability. However, considering the few cultivars with available ESTs, these results indicate at least one cysteine-containing ω-gliadin is a common feature of wheat germplasm.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- BLAST:
-
Basic local alignment search tool
- cv:
-
Cultivar
- ESTs:
-
Expressed sequence tags
- HMW:
-
High molecular weight
- LMW:
-
Low molecular weight
- NCBI:
-
National Center for Biotechnology Information
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
References
Altenbach SB, Kothari KM (2007) Omega gliadin genes expressed in Triticum aestivum cv. Butte 86: effects of post-anthesis fertilizer on transcript accumulation during grain development. J Cereal Sci 46:169–177
Anderson OD (1991) Characterization of members of a pseudogene subfamily of the wheat alpha-gliadin storage protein genes. Plant Mol Biol 16:335–337
Anderson OD, Litts JC, Greene FC (1997) The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin gene genomic clones, evidence for limited sequence conservation of flanking DNA, and Southern analysis of the gene family. Theor Appl Genet 95:50–58
Anderson OD, Rausch C, Moullet O, Lagudah E (2003) The wheat D-genome HMW-glutenin locus: BAC sequencing, gene distribution, and retrotransposon clusters. Funct Integr Genomics 3:56–68
Anderson OD, Carollo V, Chao S, Laudencia-Chingcuanco D, Lazo GR (2004) The use of ESTs to analyze the spectrum of wheat seed proteins. In: The Gluten Proteins. Proceedings 8th Gluten Workshop, pp 18–21
Battais F, Courcoux P, Popineau Y, Kanny G, Moneret-Vautrin DA, Denery-Papini S (2005) Food allergy to wheat: differences in immunoglobulin E-binding proteins as a function of age or symptoms. J Cereal Sci 42:109–117
Bianco PR, Kowalczykowski SC (1997) The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc Nat Acad Sci U S A 94:6706–6711
Cenci A, Chantret N, Kong X, Gu Y-Q, Anderson OD, Fahima T, Distelfeld A, Dubcovsky J (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor Appl Genet 107:931–939
D’Ovidio R, Masci S (2004) The low-molecular-weight glutenin subunits of wheat gluten. J Cereal Sci 39:321–339
Dupont FM, Vensel WH, Chan R, Kasarda DD (2000) Characterization of the 1B-type ω-gliadins from Triticum aestivum cultivar Butte. Cereal Chem 77:607–614
Dupont FM, Vensel W, Encarnacao T, Chan R, Kasarda DD (2004) Similarities of omega gliadins from Triticum urartu to those encoded on chromosome 1A of hexaploid wheat and evidence for their post-translational processing. Theor Appl Genet 108:1299–1308
Dupont FM, Hurkman WJ, Vensel WH, Chan R, Lopez R, Tanaka CK, Altenbach SB (2006) Differential accumulation of sulfur-rich and sulfur-poor wheat flour proteins is affected by temperature and mineral nutrition during grain development. J Cereal Sci 44:101–112
Gao S, Gu YQ, Wu J, Coleman-Derr D, Huo N, Crossman C, Jia J, Zuo Q, Ren Z, Anderson OD, Kong X (2007) Rapid evolution and complex structural organization in genomics regions harboring multiple prolamine genes in the polyploidy wheat genome. Plant Mol Biol 65:189–203
Gianibelli MC, Masci S, Larroque OR, Lafiandra D, MacRitchie F (2002) Biochemical characterization of a novel polymeric protein subunit from bread wheat (Triticum aestivum L.). J Cereal Sci 35:265–276
Hassani ME, Shariflou MR, Gianibelli MC, Sharp PJ (2008) Characterization of a ω-gliadin gene in Triticum tauschii. J Cereal Sci 47:59–67
Hayashi K, Hashimoto N, Daigen M, Ashikawa I (2004) Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet 108:1212–1220
Hsia CC, Anderson OD (2001) Isolation and characterization of wheat ω-gliadin genes. Theor Appl Genet 103:37–44
Kasarda DD, Autran J-C, Lew J-L, Nimmo CC, Shewry PR (1983) N-terminal amino acid sequences of ω-gliadins and ω-secalins. Biochim Biophys Acta 747:138–150
Kong XY, Gu YQ, You FM, Dubcovsky J, Anderson OD (2004) Dynamics of the evolution of orthologous and paralogous portions of a complex locus region in two genomes of allopolyploid wheat. Plant Mol Biol 54:55–69
Masci S, Lafiandra D, Porceddu E, Lew EJL, Tao HP, Kasarda DD (1993) D-glutenin subunits: N-terminal sequences and evidence for the presence of cysteine. Cereal Chem 70:581–585
Masci S, Egorov TA, Ronchi C, Kuzmicky DD, Kasarda DD, Lafiandra D (1999) Evidence for the presence of only one cysteine residue in the D-type low molecular weight subunits of wheat glutenin. J Cereal Sci 29:17–25
Masoudi-Nejad A, Nasuda S, Kawabe A, Endo TR (2002) Molecular cloning, sequencing, and chromosome mapping of a 1A-encoded ω-type prolamin sequence from wheat. Genome 45:661–669
Matsuo H, Kohno K, Morita E (2005) Molecular cloning, recombinant expression and IgE-binding epitope of ω-gliadin, a major allergen in wheat-dependent exercise-induced anaphylaxis. FEBS J 272:4431–4438
Metakovsky EV, Akhmedov MG, Sozinov AA (1986) Genetic analysis of gliadin-encoding genes reveals clusters as well as single remote genes. Theor Appl Genet 73:278–285
Payne PI (1987) Genetics of wheat storage proteins and the effects of allelic variation on bread-making quality. Ann Rev Plant Physiol 38:141–153
Payne PI, Holt LM, Lister PG (1988) Gli-A3 and Gli-B3, two newly designated loci coding for omega-type gliadins and D subunits of glutenin. In: Miller TE, Koebner RMD (eds) Proceedings 7th International Wheat Genetics Symposium. Bath Press, Bath, pp 999–1002
Pogna NE, Metakovsky EV, Redaelli R, Raineri F, Dachkevitch T (1993) Recombination mapping of Gli-5, a new gliadin-coding locus on chromosomes 1A and 1B in common wheat. Theor Appl Genet 87:113–121
Sabelli PA, Shewry PR (1991) Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor Appl Genet 83:209–227
Seilmeier W, Valdez I, Mendez E, Wieser H (2001) Comparative investigations of gluten proteins from different wheat species. Eur Food Res Tech 212:355–363
Shewry PR, Halford NG, Lafiandra D (2003) Genetics of wheat gluten proteins. Adv Genet 49:111–184
Song R, Llaca V, Linton E, Messing J (2001) Sequence, regulation, and evolution of the maize 22-kD α zein gene family. Genome Res 11:1817–1825
Tatham AS, Shewry PR (1995) The S-poor prolamins of wheat, barley, and rye. J Cereal Sci 22:1–16
Wang A, Gao L, Li X, Zhang Y, He Z, Xia X, Zhang Y, Yan Y (2008) Characterization of two 1D-encoded ω-gliadin subunits closely related to dough strength and pan bread-making quality in common wheat (Triticum aestivum L.). J Cereal Sci 47:528–535
Wieser H (2007) Chemistry of glutenin proteins. Food Microbiol 24:115–119
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1S
Repeat motifs of ω-gliadin proteins. Derived amino acid sequences of several ω-gliadin sequences are arrayed vertically to indicate repeat motif structure. Each vertical array shows the signal peptide, short non-repetitive N-terminal mature sequence, the series of repeat motifs, and the short non-repetitive C-terminal sequence. Underlined motifs in genes 1 and Ψ2 suggest how a deletion of the intervening repeats in Gene 1 could create a truncated gene such as Ψ2 (deletion site indicated by a line). A γ-gliadin (AF234646) repeat domain is included to show similarity to A/D-genome ω-gliadin repeat domains (F20b and G3). The line between the last two repeats in gene F20b is the site of additional sequence found in an AREL consensus sequence in Fig. 7 (PDF 12 kb).
Supplementary Fig. 2S
Divergence of 1B vs. 1A/1D ω-gliadins. The derived amino acid sequences of a 1A/1D type ω-gliadin (G3; AF280606), a 1B ω-gliadin (gene 1; EF426564), and a γ-gliadin (γ13; AF234646) are compared pairwise in dot plots. Matching criteria was 60% match over a window of 20 amino acids. a G3 vs. gene 1. b G3 vs. γ13. c Gene 1 vs. γ13 (PDF 17 kb).
Supplementary Fig. 3aS
Compare 1A and 1D ω-gliadin DNA sequences. All reported 1A and 1D ω-gliadin DNA sequences are aligned with Clustal V. Names are by Genbank accession, gene name (if named), and first author of their report (if published). Base differences are indicated by background colors, i.e., yellow if only a single difference within the set of sequences, blue and green background if two or three differences exist, respectively. Fig. 3bS is a continuation of the sequence alignment from Fig. 3aS (PDF 16 kb).
Supplementary Fig. 3bS
(PDF 16 kb)
Supplementary Fig. 4S
Primer pairs. a Shown are the primer names, primer sequences, target template, and DNA sources for primers used to PCR amplify specific ω-gliadin sequences. b Positions of primer pairs on example members of gene clusters (PDF 19 kb).
Supplementary Fig. 5S
AREL and ARQL consensus DNA sequences (PDF 6 kb).
Supplementary Table 1S
ESTs in sequence assemblies. Listed are the ESTs whose derived amino acid sequences were used in ω-gliadin sequence assemblies for the AREL and ARQL consensus sequences and 1B sequences from specific cultivars (PDF 69 kb).
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Anderson, O.D., Gu, Y.Q., Kong, X. et al. The wheat ω-gliadin genes: structure and EST analysis. Funct Integr Genomics 9, 397–410 (2009). https://doi.org/10.1007/s10142-009-0122-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-009-0122-2