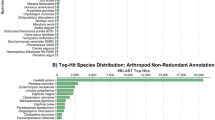
Abstract
Echinolittorina snails inhabit the upper intertidal rocky shore and face strong selection pressures from thermal extremes and fluctuations. Revealing the molecular processes of adaptive significance is greatly obstructed by the scarcity of genomic resource for these taxa. Here, we reported the first comprehensive transcriptome dataset for the genus Echinolittorina. Using Illumina HiSeq 2000 platform, about 52 M and 54 M paired-end clean reads were, respectively, generated for the control and heat-stressed libraries. Totally, 115,211 unique transcript fragments (unigenes) were assembled, with an average length of 453 bp and a N50 size of 492 bp. Approximately one third of the unigenes could be annotated according to their homology matches against the Nr, Swiss-Prot, COG, or KEGG databases, and they were found to represent 23,098 non-redundant genes. Gene expression comparison revealed that 1,267 and 6,663 annotated genes were, respectively, up- and downregulated with at least twofold changes upon heat stress. Gene Ontology and KEGG pathway analyses indicated that there were overrepresented amount of genes enriched in a broad spectrum of biological processes and pathways, including those associated with cytoskeleton organization, developmental regulation, signaling transduction, infection, and cardiac function. In addition, a transcriptome-wide search for polymorphic loci yielded a total of 11,228 simple sequence repeats (SSRs) from 9,938 unigenes and 138,631 single nucleotide polymorphism (SNP) and insertion/deletion (INDEL) sites among 22,770 unigenes. The large number of transcript sequences acquired, the biological pathways identified, and the candidate microsatellite and SNP/INDEL loci discovered in the study will serve as valuable resources for further investigations of genetic differentiation and thermal adaptation among populations.



Similar content being viewed by others
References
Amaral PP, Dinger ME, Mattick JS (2013) Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics 12:254–278
Basirico L, Morera P, Primi V, Lacetera N, Nardone A, Bernabucci U (2011) Cellular thermotolerance is associated with heat shock protein 70.1 genetic polymorphisms in Holstein lactating cows. Cell Stress Chaperones 16:441–448
Boutet I, Tanguy A, Le Guen D, Piccino P, Hourdez S, Legendre P, Jollivet D (2009) Global depression in gene expression as a response to rapid thermal changes in vent mussels. Proc Biol Sci 276:3071–3079
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dunphy BJ, Ragg NL, Collings MG (2013) Latitudinal comparison of thermotolerance and HSP70 production in F2 larvae of the greenshell mussel (Perna canaliculus). J Exp Biol 216:1202–1209
Evans TG, Hofmann GE (2012) Defining the limits of physiological plasticity: how gene expression can assess and predict the consequences of ocean change. Phil Trans R Soc B 367:1733–1745
Feder ME, Walser JC (2005) The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol 18:901–910
Franks SJ, Hoffmann AA (2012) Genetics of climate change adaptation. Annu Rev Genet 46:185–208
Fu N, Wang Q, Shen HL (2013) De novo assembly, gene annotation and marker development using Illumina paired-end transcriptome sequences in celery (Apium graveolens L.). PLoS One 8:e57686
Galindo J, Moran P, Rolan-Alvarez E (2009) Comparing geographical genetic differentiation between candidate and noncandidate loci for adaptation strengthens support for parallel ecological divergence in the marine snail Littorina saxatilis. Mol Ecol 18:919–930
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Gracey AY, Chaney ML, Boomhower JP, Tyburczy WR, Connor K, Somero GN (2008) Rhythms of gene expression in a fluctuating intertidal environment. Curr Biol 18:1501–1507
Hammond LM, Hofmann GE (2010) Thermal tolerance of Strongylocentrotus purpuratus early life history stages: mortality, stress-induced gene expression and biogeographic patterns. Mar Biol 157:2677–2687
Helmuth B, Harley CD, Halpin PM, O’donnell M, Hofmann GE, Blanchette CA (2002) Climate change and latitudinal patterns of intertidal thermal stress. Science 298:1015–1017
Henderson B (2010) Integrating the cell stress response: a new view of molecular chaperones as immunological and physiological homeostatic regulators. Cell Biochem Funct 28:1–14
Hofmann GE, Todgham AE (2010) Living in the now: physiological mechanisms to tolerate a rapidly changing environment. Annu Rev Physiol 72:127–145
Hollenbeck CM, Portnoy DS, Gold JR (2012) Use of comparative genomics to develop EST-SSRs for red drum (Sciaenops ocellatus). Mar Biotechnol (NY) 14:672–680
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol :138–148
Ivanina AV, Taylor C, Sokolova IM (2009) Effects of elevated temperature and cadmium exposure on stress protein response in eastern oysters Crassostrea virginica (Gmelin). Aquat Toxicol 91:245–254
Kultz D (2005) Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol 67:225–257
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Leung AK, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40:205–215
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAM tools. Bioinformatics 25:2078–2079
Liu WG, Huang XD, Lin JS, He MX (2012) Seawater acidification and elevated temperature affect gene expression patterns of the pearl oyster Pinctada fucata. PLoS One 7:e33679
Liu S, Wang X, Sun F, Zhang J, Feng J, Liu H, Rajendran KV, Sun L, Zhang Y, Jiang Y, Peatman E, Kaltenboeck L, Kucuktas H, Liu Z (2013) RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol Genomics 45:462–476
Lockwood BL, Somero GN (2012) Functional determinants of temperature adaptation in enzymes of cold- versus warm-adapted mussels (Genus Mytilus). Mol Biol Evol 29:3061–3070
Lockwood BL, Sanders JG, Somero GN (2010) Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J Exp Biol 213:3548–3558
Maron BJ (2002) Hypertrophic cardiomyopathy: a systematic review. JAMA 287:1308–1320
Marshall DJ, McQuaid CD (2011) Warming reduces metabolic rate in marine snails: adaptation to fluctuating high temperatures challenges the metabolic theory of ecology. Proc Biol Sci 278:281–288
Marshall DJ, Dong YW, McQuaid CD, Williams GA (2011) Thermal adaptation in the intertidal snail Echinolittorina malaccana contradicts current theory by revealing the crucial roles of resting metabolism. J Exp Biol 214:3649–3657
Martinez-Fernandez M, Bernatchez L, Rolan-Alvarez E, Quesada H (2010) Insights into the role of differential gene expression on the ecological adaptation of the snail Littorina saxatilis. BMC Evol Biol 10:356
Micallef G, Bickerdike R, Reiff C, Fernandes JMO, Bowman AS, Martin SM (2012) Exploring the transcriptome of Atlantic salmon (Salmo salar) skin, a major defense organ. Mar Biotechnol (NY) 14:559–569
Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F (2013) Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol 24:22–30
Negri A, Oliveri C, Sforzini S, Mignione F, Viarengo A, Banni M (2013) Transcriptional response of the mussel Mytilus galloprovincialis (Lam.) following exposure to heat stress and copper. PLoS One 8:e66802
Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A (2008) Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 180:1065–1071
Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance—degradation and reactivation of damaged proteins. Annu Rev Genet 27:437–496
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Podrabsky JE, Somero GN (2004) Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J Exp Biol 207:2237–2254
Portner HO (2010) Oxygen- and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J Exp Biol 213:881–893
Posner M, Kiss AJ, Skiba J, Drossman A, Dolinska MB, Hejtmancik JF, Sergeev YV (2012) Functional validation of hydrophobic adaptation to physiological temperature in the small heat shock protein αA-crystallin. PLoS One 7:e34438
Rebl A, Verleih M, Kobis JM, Kuhn C, Wimmers K, Kollner B, Goldammer T (2013) Transcriptome profiling of gill tissue in regionally bred and globally farmed rainbow trout strains reveals different strategies for coping with thermal stress. Mar Biotechnol (NY) 15:445–460
Reid DG, Lal K, Mackenzie-Dodds J, Kaligis F, Littlewood DTJ, Williams ST (2006) Comparative phylogeography and species boundaries in Echinolittorina snails in the central Indo-West Pacific. J Biogeogr 33:990–1006
Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40:253–266
Robinson MD, Mccarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Robinson N, Sahoo PK, Baranski M, Das Mahapatra K, Saha JN, Das S, Mishra Y, Das P, Barman HK, Eknath AE (2012) Expressed sequences and polymorphisms in rohu carp (Labeo rohita, Hamilton) revealed by mRNA-seq. Mar Biotechnol (NY) 14:620–633
Ronges D, Walsh JP, Sinclair BJ, Stillman JH (2012) Changes in extreme cold tolerance, membrane composition and cardiac transcriptome during the first day of thermal acclimation in the porcelain crab Petrolisthes cinctipes. J Exp Biol 215:1824–1836
Runcie DE, Garfield DA, Babbitt CC, Wygoda JA, Mukherjee S, Wray GA (2012) Genetics of gene expression responses to temperature stress in a sea urchin gene network. Mol Ecol 21:4547–4562
Sanford E, Kelly MW (2011) Local adaptation in marine invertebrates. Annu Rev Mar Sci 3:509–535
Schoville SD, Barreto FS, Moy GW, Wolff A, Burton RS (2012) Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol Biol 12:170
Smith S, Bernatchez L, Beheregaray LB (2013) RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genomics 14:375
Somero GN (2010) The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J Exp Biol 213:912–920
Sorensen JG, Nielsen MM, Kruhoffer M, Justesen J, Loeschcke V (2005) Full genome gene expression analysis of the heat stress response, in Drosophila melanogaster. Cell Stress Chaperones 10:312–328
Stenseng E, Braby CE, Somero GN (2005) Evolutionary and acclimation-induced variation in the thermal limits of heart function in congeneric marine snails (genus Tegula): implications for vertical zonation. Biol Bull 208:138–144
Stillman JH, Tagmount A (2009) Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol Ecol 18:4206–4226
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L). Theor Appl Genet 106:411–422
Thomas JO, Travers AA (2001) HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26:167–174
Tomanek L (2010) Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. J Exp Biol 213:971–979
Truebano M, Burns G, Thorne MS, Hillyard G, Peck LS, Skibinski DOF, Clark MS (2010) Transcriptional response to heat stress in the Antarctic bivalve Laternula elliptica. J Exp Mar Biol Ecol 391:65–72
Vandenkoornhuyse P, Dufresne A, Quaiser A, Gouesbet G, Binet F, Francez AJ, Mahe S, Bormans M, Lagadeuc Y, Couee I (2010) Integration of molecular functions at the ecosystemic level: breakthroughs and future goals of environmental genomics and post-genomics. Ecol Lett 13:776–791
Verghese J, Abrams J, Wang Y, Morano KA (2012) Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76:115–158
Walters RJ, Blanckenhorn WU, Berger D, Woods A (2012) Forecasting extinction risk of ectotherms under climate warming: an evolutionary perspective. Funct Ecol 26:1324–1338
Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H (2011) SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res 39:e132
Werner GD, Gemmell P, Grosser S, Hamer R, Shimeld SM (2013) Analysis of a deep transcriptome from the mantle tissue of Patella vulgata Linnaeus (Mollusca: Gastropoda: Patellidae) reveals candidate biomineralising genes. Mar Biotechnol (NY) 15:230–243
Whitehead A (2012) Comparative genomics in ecological physiology: toward a more nuanced understanding of acclimation and adaptation. J Exp Biol 215:884–891
Zuo W, Moses ME, West GB, Hou C, Brown JH (2012) A general model for effects of temperature on ectotherm ontogenetic growth and development. Proc Biol Sci 279:1840–1846
Acknowledgments
We are grateful to two anonymous reviewers for their comments on an earlier version of this manuscript. The authors thank Aldrin KY Yim for the access of server at the Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong (CUHK) and Long Fan for his assistance in compiling the PERL scripts to process the data. The project was supported by a grant of the Group Research Scheme from the Research Committee, CUHK.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Figure S1
E-value and similarity distribution in Nr annotation. (A) 54.6 % of the annotated unigenes displayed strong homology (e-value smaller than 1.0E-15) to known genes, whereas 45.4 % of the sequences have e-values ranging from 1.0E-15 to 1.0E-5. (B) 63.9 % of the unigenes showed a similarity higher than 40 %, and 36.1 % of the sequences had a similarity ranging from 40 % to 15 %. (JPEG 77 kb)
Table S1
Statistics of functional annotation. The number of unigenes annotated to each database is shown. (PDF 80 kb)
Table S2
Annotation of the assembled sequences. The unigenes were analyzed for similarities with sequences recorded in major databases, including the nr, Swiss-Pro, COG and KEGG databases. (XLSX 14567 kb)
Table S3
KEGG pathways present in the snail transcriptome. A total of 23,016 unigenes could find homology hits in the KEGG database. These unigenes represent 258 predicted KEGG pathways. The number of unigenes in each pathway is summarized. (XLSX 20 kb)
Table S4
List of unigenes differentially expressed upon heat stress. A total of 8,304 transcript-derived unigenes were detected as up-regulated while 22,359 unigenes were down-regulated. Annotation of these unigenes to the nr database is provided. (XLSX 3456 kb)
Table S5
List of over-represented KEGG pathways. 51 KEGG pathways were found significantly over-represented in the heat-stressed animals. For each pathway, the number of unigenes differentially expressed, the total number of unigenes, the corrected p-value and pathway ID are shown. (PDF 9 kb)
Table S6
List of SNP/INDEL loci identified in the assembled unigenes. A total of 138,631 candidate SNP/INDEL loci were identified among 22,770 unigenes. (XLSX 11143 kb)
Table S7
List of protein-coding unigenes with SNP/INDEL sites. 8,590 unigenes with SNP/INDEL sites corresponded to protein-coding genes. Annotation of these unigenes is provided. (XLSX 774 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Hui, J.H.L., Chan, T.F. et al. De Novo Transcriptome Sequencing of the Snail Echinolittorina malaccana: Identification of Genes Responsive to Thermal Stress and Development of Genetic Markers for Population Studies. Mar Biotechnol 16, 547–559 (2014). https://doi.org/10.1007/s10126-014-9573-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-014-9573-0