Abstract
Background
Increasing evidence indicates that leucine-rich-alpha-2-glycoprotein 1 (LRG1) is associated with multiple malignancies, but whether it participates in gastric cancer (GC) angiogenesis remains unclear.
Methods
The expression levels of LRG1 were assessed in GC samples. Endothelial tube formation analysis, HUVEC migration assay, chorioallantoic membrane assay (CAM), and xenograft tumor model were used to investigate the effect of LRG1 on angiogenesis in gastric cancer. The involvement of activating transcription factor 3 (ATF3) was analyzed by chromatin immunoprecipitation (ChIP) and dual-luciferase reporter assay. Western blot and enzyme-linked immunosorbent assay were performed to measure the SRC/STAT3/VEGFA pathway.
Results
LRG1 was overexpressed in GC tissues and associated with cancer angiogenesis. In addition, LRG1 markedly promoted GC cell proliferation in vitro and in vivo. Moreover, overexpression of LRG1 could stimulate GC angiogenesis in vitro and in vivo. Then, we identified ATF3 promotes the transcription of LRG1 and is a positive regulator of angiogenesis. Additionally, LRG1 could activate VEGFA expression via the SRC/STAT3/ VEGFA pathway in GC cells, thus contributing to the angiogenesis of GC.
Conclusions
The present study suggests LRG1 plays a crucial role in the regulation of angiogenesis in GC and could be a potential therapeutic target for GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is the fifth most diagnosed malignancy worldwide with over 1 million estimated new cases annually. Since gastric cancer is often diagnosed at an advanced stage, its mortality rate is high, making it the fourth most common cause of cancer-related death [1, 2]. Although early gastric cancer can be resected for therapeutic purposes, treatment options are limited in cases of advanced metastatic or unresectable tumors [3, 4]. Combination chemotherapy can sometimes turn initially unresectable stomach cancer into resectable cancer [5, 6]. The application of antiangiogenic drugs in the treatment of gastric cancer has attracted more attention [7, 8]. Therefore, clarifying the molecular events related to gastric cancer and finding new biomarkers and therapeutic targets are indispensable and urgent for the clinical outcome of gastric cancer.
Leucine-rich-alpha-2-glycoprotein1(LRG1) was firstly isolated and characterized from human serum by Haupt and Baudner in 1977 [9]. It is a secreted glycoprotein with an amino acid sequence divided into 13 segments of 24 residues each, 8 of which exhibit a periodic pattern of repeating consensus sequences, which enables its interaction with other proteins [10]. Previous studies have demonstrated that LRG1 is widely involved in signal transduction, immune response and cell proliferation, cell migration, cell invasion, cell adhesion, cell apoptosis, and neovascularization [11,12,13,14].
LRG1 has been reported to be associated with the pathogenesis of a variety of diseases, including cancer [15] and inflammatory diseases [11, 16]. LRG1 is overexpressed in several types of malignant tumors, including pancreatic [17], colon [18], lung [19], bladder [20], ovarian [21], and endometrial [22] cancer. LRG1 binds directly to TGF-β accessory receptor endoglin and enhances the activation of downstream signaling pathways, including Smad1/5/8, resulting in the promotion of angiogenesis in endothelial cells [13]. It was reported that patients with gastric cancer with high LRG1 expression had a poorer prognosis, and serum LRG1 may be a useful prognostic marker in gastric cancer patients [23]. However, the role of LRG1 in the regulation of angiogenesis of GC remains functionally uncharacterized.
In the present study, we identify that LRG1 is overexpressed in GC tumor tissues and cell lines. LRG1 overexpression in GC cells stimulated proliferation and angiogenesis both in vitro and in vivo. ATF3 promotes the transcription of LRG1 in GC cells. LRG1 promotes angiogenesis in GC tumors through modulation of the SRC/STAT3/VEGFA pathway.
Materials and methods
Cell lines and cell culture
The human gastric cancer cell lines SGC7901 were purchased from American Type Culture Collection (ATCC). AGS, MFC, MKN45, MGC803, and GES-1 cell lines were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. All GC cell lines were identified by short tandem repeat analysis, and the results of mycoplasma test were negative. AGS, MFC, MKN45, and SGC7901 were cultured with RPMI-1640 medium (Gibco, San Francisco, CA, USA) containing 10% fetal bovine serum (Gibco) with 100 U/mL penicillin and streptomycin (Servicebio, Wuhan, China) at 37 ℃ in a humidified incubator of 5% CO2. MGC803 and GES-1 cell lines were cultured with DMEM medium (Gibco) containing10% fetal bovine serum with 100 U/mL penicillin and streptomycin.
HUVEC extraction and identification
Human umbilical vein endothelial cells (HUVECs) were obtained from human umbilical veins using Type I collagenase Hank’s solution. HUVECs were cultured in Endothelial Cell Medium (ECM) (cat. No.1001, ScienceCell, San Diego, California, USA) containing 25 ml of fetal bovine serum (FBS, Cat. No. 0025), 5 ml of endothelial cell growth supplement (ECGS, Cat. No. 1052), and 5 ml of antibiotic solution (P/S, Cat. No. 0503). Cells were identified by immunofluorescence staining of anti-von Willebrand factor (VWF) (Bs-0586R, Bioss) and anti-CD31(ab9498, Abcam) for cell surface antigens (Supplementary Fig. S1).
Animal studies and clinical specimens
BALB/c nude mice were purchased from the Beijing Huafukang Biological Co., Ltd and housed under standard pathogen-free conditions in the Experimental Animal Center of Tongji Medical College, Huazhong University of Science & Technology. The human sample was from the Endoscopy Center of Union Hospital. All samples were obtained with the patients’ informed consent, and the samples were processed histologically.
Virus construction and infection
Lentivirus for LRG1 overexpression and shRNA sequences targeting LRG1 (shRNA, shLRG1 #1 and shLRG1 #2) was designed and constructed by Obio Technology Co. Ltd., Shanghai, China. Lentivirus was added to the culture medium of GC cells by using HitransG A (Genechem, Shanghai, China) and puromycin for screening stable clones following the manufacturer’s recommendations. The targeted LRG1 sequences were as follows: CATGCTGGACCTCTCCAATAA (shLRG1 #1) and TGGACACCCTGGTATTGAAAG (shLRG1 #2). The amplification primers for the LRG1 coding region were as follows: GGCTGAAGCTTGCAGAGCTACCATGTCCTC.
RNA extraction and real-time qPCR
Total RNA was extracted from samples with RNAiso Plus reagent (TAKARA, Japan) and quantified by using Nanodrop 8000 and stored at − 80 °C. 1000 ng of RNA was reversely transcribed into cDNA using a reverse transcription system (TAKARA, Japan). Real-Time qPCR was performed to quantify the transcripts using TB Green PCR Master Mix (TAKARA, Japan). The primers for the PCR reaction are shown in Supplementary Table S1. The relative abundance of RNA was normalized to GAPDH. 2−(ΔΔCt) was used to calculate the relative abundance of mRNA.
In vitro proliferation assay
CCK-8 cell proliferation assay: cells were seeded in 96-well plates at 1000 cells/well. Cell proliferation was detected using CCK-8 kit at 450 nm following manufacturer’s instructions.
Colony formation assay: GC cells (1 × 104 cells) were plated into each well of a 12-well plate and cultured at 37 °C for 10 days, with addition of fresh growth medium every 3 days. The colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet solution for 20 min. Then the colonies were counted.
In vitro migration assay
1000 HUVEC cells were suspended in serum-free medium and added to the upper chamber of the 24-well insert (membrane pore size, 3 mm; Corning, USA). Medium supplemented with 10% serum was added to the lower chamber. After incubation for 18 h at 37° in an incubator, unmigrated cells on the top membrane were removed with cotton swabs. Cells that migrated were fixed in 4% paraformaldehyde for 20 min and stained with 1% crystal violet for 10 min. Cells were counted in five different fields using an inverted light microscope.
Endothelial cell tube formation assay
10 μl growth factor reduced Matrigel (Corning, BD Biosciences, CA, USA) was pre-coated into a μ-Slide Angiogenesis plate (ibidi, Martin Reid, Germany) and polymerized at 37 °C for 30 min. 1 × 104 HUVECs pretreated with calcein AM were resuspended and seeded into the μ-Slide Angiogenesis plate. After 7 h of incubation with conditioned ECM medium with tumor supernatants, capillary-like tubes were photographed using a confocal microscope. The mesh number of tubular structures was quantified by the NIH Image J. Data were obtained from three independent experiments.
Chick embryo chorioallantoic membrane (CAM) assay
The CAM assay was performed as previously described [24]. Briefly, after cutting a small puncture window, 30 μl of cell suspension was dropped into a gelatin sponge and added onto the CAM in each 8-day-old fertilized egg. On day 12, the eggs were opened, and the CAM vasculatures were imaged using a stereomicroscope equipped with a digital camera. Angiogenesis was quantified by determining the blood vessel density (percentage of blood vessel area over the whole area under a microscopic field) using the image analysis program IPP 6.0 (Image-Pro Plus, version 6.0, Media Cybernetics).
In vivo tumorigenesis
BALB/c nude mice (4-week-old) were obtained from Beijing Huafukang Biological Co., Ltd and housed under standard pathogen-free conditions in Experimental Animal Center of Tongji Medical College, Huazhong University of Science & Technology. 2 × 106 gastric cancer cells were injected into the right upper flanks of mice. The tumor volumes were measured and calculated every 2 days by the formula (width2 × length)/2. All mice were killed after 21 days and the subcutaneous tumor was removed, imaged, and placed in 4% formaldehyde for further experiments.
Chromatin immunoprecipitation (ChIP)
For ChIP assays, a ChIP kit (Millipore) protocol was performed. SGC7901 and MFC cells were used for each immunoprecipitation. The protein and DNA were cross-linked with 1% formaldehyde, cleaved with SDS buffer, and cut by ultrasound. One ATF3 antibody (Abcam, ab207434) was used for immunoprecipitation. Next, ChIP qRT-PCR was performed to validate the pulled samples. ChIP qRT-PCR primer sequences are listed in Supplementary Table S2.
Luciferase reporter assay
The LRG1 promoter was cloned from human genomic cDNA using Q5 Hotstart Polymerase and inserted into the pGL4.10 Basic vector (pGL4-LRG1, pGL4-LRG1 -del A, pGL4-LRG1-del B, pGL4-LRG1-del C). Cells were co-transfected with plasmid and siRNA using Lipofectamine 2000 (Invitrogen) and plated in a well of a 96-well plate. Luciferase reporter assay was performed by using a dual-luciferase assay (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Immunohistochemistry assay
Immunohistochemistry analysis was conducted as previously described. In brief, all paraffin embedded material was sectioned at 4 μm. After dewaxing and hydration, the sections were incubated overnight with antibodies against human LRG1(ab178698, Abcam), human PECAM1 (ab9498, Abcam), mouse CD31 (AF3628-SP, R&D Systems), mouse Ki-67 (ab1667, Abcam), and human ATF3 (ab254268, Abcam). Primary antibody was detected with horseradish peroxidase-conjugated secondary antibodies incubated for 8 min. Sections were washed in distilled water, counterstained with hematoxylin, dehydrated and mounted. The whole tissue section was scored with staining intensity and percentage and the scoring scale was graded as follows: 0 points (no staining), 1 point (light brown staining), 2 points (brown staining) and 3 points (dark brown staining). The percentage of positive cells is divided into four levels: 1 point (< 5%), 2 points (5–30%), 3 points (31–60%), 4 points (61–100%). The IHC staining score was calculated as follows: intensity score × percentage score.
Western blotting
Cells were lysed in cold RIPA buffer containing protease inhibitors. Equal amounts of total protein were separated by SDS-PAGE (One-Step SDS-PAGE gel fast preparation kit, Biotides, China), transferred to PVDF membranes, and blocked in 5% milk. The primary antibodies were diluted according to the instructions. Membranes were incubated overnight with primary antibodies at 4 °C, followed by secondary antibodies for 1 h at room temperature. Blots were developed using Millipore Immobilon Western Chemiluminescent HRP Substrate.
Statistical analysis
All experiments were repeated at least three times, and the results were expressed as means ± SD. Student’s t test or the one-way analysis of variance (ANOVA) were used to analyze the data, and Chi-square test was used to analyze differences in other variables, as appropriate. All statistical analyses were performed using GraphPad Prism software. p value of < 0.05 was considered statistically significant for all datasets (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
LRG1 was overexpressed in human GC and predicts poor survival
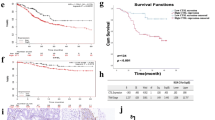
Previous studies have demonstrated that LRG1 was overexpressed in a wide range of human malignancies, including pancreatic [17], colon [18], and lung [19] cancers. We performed bioinformatic analysis of LRG1 expression profiles through the Cancer Genome Atlas (TCGA) and found that LRG1 was overexpressed in breast carcinoma, glioblastoma multiforme, kidney renal clear cell carcinoma, lung adenocarcinoma, stomach adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma. (Fig. 1A). Notably, LRG1 was obviously upregulated in GC tissues (Fig. 1B). Moreover, we identified significantly increased LRG1 mRNA expression levels in gastric tumors relative to those in normal gastric tissues (Fig. 1C). LRG1 protein expressions were examined and significantly higher expression levels of LRG1 were detected in GC tissues than in corresponding normal tissues (Fig. 1D, E). We then examined LRG1 protein expression in GC tissues and corresponding non-tumor tissues by IHC. The IHC results showed that the average expression score of LRG1 was significantly higher in GC tissues than in adjacent non-tumor tissues (Fig. 1F, G). In addition, Kaplan–Meier Plotter analysis showed that LRG1 expression was negatively correlated with the survival of GC patients (Fig. 1H). LRG1 expression has been demonstrated to be associated with angiogenesis [13]. IHC of staining of PECAM1 was performed to examine the blood vessel density (VD) and the results showed that VD in GC tumor tissues was significantly higher than that that of normal gastric mucosa (Fig. 1I, J). LRG1 expression was positively correlated with the VD of GC tissues (Fig. 1K).
Overexpression of LRG1 is correlated with vessel density in human GC tissues and predicts a poor prognosis. A The expression of LRG1 in a pan-cancer dataset. B Overexpression of LRG1 mRNA in GC tissues than normal tissues in the TCGA dataset. C LRG1 expression level in GC tissues and adjacent non-tumor tissues detected by qRT-PCR. D LRG1 expression level in GC tissues and adjacent non-tumor tissues was detected by western blot. E Quantification of (D). ***p < 0.001. F Representative IHC staining for LRG1 in GC and adjacent non-tumor tissues. G Quantification of (F). ***p < 0.001. H The overall survival analysis was plotted using Kaplan–Meier Plotter for patients with GC. I Representative IHC staining for PECAM1 in GC and adjacent non-tumor tissues. J Quantification of (I). ***p < 0.001. K Correlation analysis of LRG1 and vessel density in GC
LRG1 promotes GC cell proliferation in vitro and in vivo
To identify the functional role of LRG1 in GC cells, we evaluated the expression of LRG1 in GC cell lines via qRT-PCR and western blotting (Fig. 2A, B). LRG1 knockdown by lentivirus-mediated shRNA or overexpressing experiments were performed. Real-time PCR and western blotting were performed to detect whether the expression level of LRG1 changed in GC cells (Fig. 2C, D). Furthermore, two-dimensional (2D) colony formation assay and CCK-8 assay demonstrated that LRG1 knockdown significantly inhibited the proliferation of SGC7901 and AGS cells. In contrast, overexpression of LRG1 could significantly promote MGC803 cell proliferation (Fig. 2E, F). Given the in vitro findings described above, a nude mice xenograft model was established to investigate the effect of LRG1 knockdown in vivo. The results showed that the tumors generated by SGC7901/shLRG1 cells exhibited smaller volume and size, and less weight than those generated by SGC7901/shNC cells (Fig. 2G–J). HE staining and IHC of Ki67 showed reduced numbers of proliferating cells in SGC7901/shLRG1 tumors compared to SGC7901/shNC tumors (Fig. 2K–L). Thus, these findings suggest that LRG1 in GC cells plays an oncogenic role in gastric cancer.
LRG1 stimulates GC cells proliferation ability in vitro and in vivo. A, B Expression of LRG1 gene and protein in GC cell lines and a normal gastric epithelium cell line. C, D qRT-PCR and western blot analysis of LRG1 expression in GC cells after shRNA knockdown or overexpression of LRG1. E Colony formation assay of indicated GC cell lines. **p < 0.01. F CCK-8 assay of indicated GC cell lines. G–H Images of subcutaneous tumors formed by SGC7901/shNC and SGC7901/shLRG1 cells. I Growth curve of tumors described in (G). J Average tumor weights from nude mice. K H&E staining and Ki-67 immunostaining of tumors described in (G). L Quantification of Ki-67 staining. **p < 0.01
LRG1 enhances endothelial cells proliferation, migration, and tube formation
Given that LRG1 was reported to promote angiogenesis [13], we determined whether knockdown or ectopic overexpression of LRG1 in GC cells could affect tube formation and migration of endothelial cells. Conditioned medium (CM) collected from LRG1-modified GC sublines was added to a 3D culture of human umbilical vein endothelial cells (HUVECs). In tube formation assays, the mesh number of tubular structures formed by HUVECs in CM from SGC7901/shLRG1 cells was significantly decreased compared with CM from the SGC7901/shNC cells (Fig. 3A, B), whereas CM from MGC803/LRG1 cells elicited more tube formation by HUVECs in comparison with the control group (MGC803/vector) (Fig. 3C, D). Tumor cells may regulate endothelial cell tube formation by altering their proliferation and migration. To examine these two possibilities, we performed Transwell assay and CCK-8 viability assay. The results showed that downregulation of LRG1 reduced the migratory ability and proliferation induced by CM from GC cells (Fig. 3E, F , I), whereas overexpression of LRG1 in GC cells promoted cell viability and migration of HUVECs (Fig. 3G, H, J). These results indicate that LRG1 could promote the proliferation, migration and tube formation of HUVEC cells in vitro.
LRG1 stimulates angiogenesis in vitro. a In vitro tube formation to investigate the effect of CM from GC cells transduced with shLRG1 or shNC. B Quantification of (A). *p < 0.05, **p < 0.01, ***p < 0.001. C In vitro tube formation to investigate the effect of CM from GC cells stably overexpressing LRG1 or control vector. D Quantification of (C). **p < 0.01. E, F Transwell assay on HUVECs to investigate the effect of CM from GC cells transduced with shLRG1 or shNC. Data shown is representative of 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. G, H Transwell assay on HUVECs to investigate the effect of CM from GC cells stably overexpressing LRG1 or control vector. Data shown is representative of three independent experiments. **p < 0.01. I CCK-8 assay of HUVECs GC cells transduced with shLRG1 or shNC. *p < 0.05, **p < 0.01. J CCK-8 assay of GC cells stably overexpressing LRG1 or control vector. **p < 0.01
LRG1 promotes GC angiogenesis in vivo
To determine the effect of LRG1 on angiogenesis in vivo, we used a chick embryo chorioallantoic membrane (CAM) assay, which has emerged as a standard for in vivo angiogenesis assay [25]. SGC7901/shLRG1 cells significantly reduced the vessel number compared to SGC7901/shNC cells (Fig. 4A, B), whereas MGC803/LRG1 cells induced the formation of more vessels than MGC803/vector cells (Fig. 4C, D). Furthermore, vascellum density in subcutaneous tumors formed by SGC7901/shNC and SGC7901/shLRG1 cells was examined by IHC of CD31 and in vivo tumor vascular permeability fluorescence imaging. Knockdown of LRG1 led to a lower vascellum density in the tumors (Fig. 4E, H). Thus, these findings suggest that LRG1 in GC cells enhance tumor progression by promoting the angiogenesis of GC.
LRG1 promotes angiogenesis in vivo. A, B Representative images and quantification of the CAM assay injected with GC cells transduced with shLRG1 or shNC. *p < 0.05, **p < 0.01. C, D Representative images and quantification of the CAM assay injected with GC cells stably overexpressing LRG1 or control vector. **p < 0.01. E IHC of CD31 in subcutaneous tumors formed by SGC7901/shNC or SGC7901/shLRG1. F Angiogenesis fluorescent agent AngioSense 750EX in tumors. G Quantification of (E). *p < 0.05. H Quantification of (G). **p < 0.01
ATF3 regulates LRG1 transcription
The expression of LRG1 mRNA was increased in human gastric tumor tissues, and human gastric cancer cell lines turned our attention to transcriptional control. By silicon-based analysis of transcription factors (TFs) performed with ALGGEN database, we identified a list of candidate transcriptional regulators that predict binding to the LRG1 promoter. From this list, we selected activating transcription factor 3 (ATF3) whose expression correlated with endothelial proliferation and angiogenesis [26]. Using western blotting, we found that ATF3 expression was significantly increased in SGC7901 cells (Fig. 5A). As shown in Fig. 5B, ATF3 was upregulated in GC tissues compared with non-tumor tissues. IHC analysis on GC tissues showed that LRG1 was positively correlated with ATF3 (Fig. 5C, D). Then we examined the effect of silencing ATF3 on LRG1 expression in gastric cancer cell lines. Depletion of ATF3 consistently resulted in reduction in LRG1 levels in GC cells (Supplementary Fig. S2) and concomitant decreased HUVEC migration and tube formation (Fig. 5E–I). To determine whether the direct regulation by ATF3 is a possible regulation pathway of LRG1 expression, we assessed the binding of ATF3 to the LRG1 promoter, which is predicted to contain three putative binding sites at site A (− 1152 to − 1163), site B (− 1400 to − 1411), and site C (− 277 to − 288) in JASPAR database. Dual-luciferase reporter assay confirmed the ability of ATF3 to control LRG1 expression at the promoter region containing site A (Fig. 5J, K). ChIP of ATF3 followed by quantitative PCR showed that ATF3 binds directly to site A (Fig. 5L). Collectively, our data reveal ATF3 as a key regulator of LRG1 transcriptional induction in angiogenesis of gastric cancer and an upstream contributor to gastric cancer progressive behavior.
ATF3 regulates LRG1 transcription. A Western blot of ATF3 in GES and SGC7901 cells. B Representative images of ATF3 immunohistochemistry staining in GC tissues. C IHC staining of ATF3 and LRG1 in GC tissues and non-tumor gastric tissues. D Correlation analysis of LRG1 and ATF3 in GC tissues. E Transwell assay and tube formation of HUVECs conducted with CM from SGC7901 and MFC cells stably overexpressing ATF3 or control vector and transfected with shNC or shLRG1. F–I Quantification of (E). *p < 0.05, **p < 0.01. J Schematic of the LRG1 promoter indicating the JASPAR-predicted binding site of ATF3 (Exon 1, 2 and 3). K Promoter luciferase reporter assay of three LRG1 promoter constructs to show LRG1 activity in MGC803/vector and MGC803/ATF3. L ChIP qPCR of the LRG1 promoter at the indicated A region in SGC7901 cells
SRC/STAT3/VEGFA are mediators of the pro-angiogenic effects of LRG1
LRG1 was reported to modulate the activation of critical pro-angiogenic molecules including HIF-1α and VEGFA [18]. LRG1 was found to interact with endoglin and promote pathogenic angiogenesis via promoting the TGF-β1/ALK1 signaling pathway [13]. Studies have shown that LRG1 gene expression was also related to signal transduction of the JAK–STAT pathway [27]. To further explore the underlying mechanism of how LRG1 promoted the angiogenesis of GC, we analyzed the activation of the downstream mediators of these signaling pathways. Western blot analysis showed that p-SRC, p-STAT3, and VEGFA levels were higher in GC than non-tumor tissues with high ATF3 and LRG1 expression (Fig. 6A). LRG1 overexpression significantly upregulated the nonreceptor tyrosine kinase SRC and STAT3 in GC cells. (Fig. 6B–D). Our data implied that LRG1 overexpression increased VEGFA mRNA expression and protein secretion level in both MGC803 and MKN45 cells (Fig. 6E, F). Similarly, human rLRG1 also increased VEGFA in GC cells in a time-dependent manner (Fig. 6G, H). To confirm whether the pro-angiogenic effects of LRG1 was dependent on the activation of SRC/STAT3 signaling pathway, GC cells were pretreated with STATTIC, a selective STAT3 inhibitor. The results showed that the promoting effect of LRG1 on HUVEC cell proliferation and migration was partially blocked after treatment of STATTIC (Fig. 6J–R). Western blot showed that a similar effect was observed by the relative expression of phosphorylation of STAT3 (Fig. 6I). Thus, LRG1 might promote GC angiogenesis through the SRC /STAT3/VEGFA signaling pathway.
SRC/STAT3/VEGFA mediates the pro-angiogenic effects of LRG1. A Activated SRC and STAT3 and VEGFA were detected in GC tissues and non-tumor tissues. B The protein levels of Src, p-Src, STAT3, and p-STAT3 expressions in response to LRG1 knockdown in SGC7901 and MGC803/ATF3 cells were measured by western blot. C, D Quantification of (B). **p < 0.01. E, F MGC803 and MKN45 cells were overexpressed with LRG1 stably, and VEGFA expression was quantified by qRT-PCR and ELISA. G, H MGC803 and MKN45 cells were conducted with rLRG1 (250 ng/ml) for 24 h and VEGFA expression was quantified by qRT-PCR and ELISA. I Western blot analysis showed that inhibiting STAT3 alleviated p-Src, p-STAT3 and VEGFA expressions in SGC7901 and MGC803/LRG1 cells. J–L Quantification of (I). **p < 0.01. M SGC7901 and MGC803/LRG1 cells were pretreated with STATTIC and VEGFA expression was quantified by ELISA. N Transwell migration assay to investigate the effect of STAT3 inhibition on HUVEC migration after treatment with CM from SGC7901 and MGC803/LRG1 cells. O Quantification of (N). **p < 0.01. P In vitro tube formation to investigate the effect of STAT3 inhibition on HUVEC tube formation after treatment with CM from SGC7901 and MGC803/LRG1 cells. Q Quantification of (P). *p < 0.05. R CCK-8 assay to investigate the effect of STAT3 inhibition on HUVEC proliferation after treatment with CM from SGC7901 and MGC803/LRG1 cells. *p < 0.05
Discussion
Gastric cancer is a globally important disease. Since gastric cancer is often diagnosed at an advanced stage, the mortality rate is high [1]. Surgical or endoscopic resection is the main treatment [28, 29]. In addition, chemotherapy improves survival and quality of life in patients with locally advanced, unresectable, or metastatic gastric cancer [30]. At present, there is no effective biomarker to guide the choice of chemotherapy for advanced gastric cancer. Therefore, it is urgent to find the target proteins or relevant signaling pathways and elucidate the exact mechanism of the occurrence and development of gastric cancer.
It was reported that patients with gastric cancer with high LRG1 expression had a poorer prognosis, and serum LRG1 may be a useful prognostic marker in gastric cancer patients [23]. In our study, we demonstrated LRG1 was increased significantly in GC tissues compared with normal tissues. Furthermore, LRG1 predicts poor prognosis of GC patients. These results suggest that LRG1 may be involved in the development and progression of gastric cancer, which is consistent with published studies on colon, pancreatic cancer, and non-small cell lung cancer [17, 18, 31, 32]. Nevertheless, LRG1 is thought to be a negative regulator of tumors in esophageal squamous cell carcinoma and hepatocellular carcinoma [33, 34]. Although previous studies have shown that LRG1 is associated with a variety of malignancies, the function of LRG1 in GC has not been fully determined. Our results showed that knockdown of LRG1 significantly attenuated the proliferation of GC cells in vitro and in vivo, which are in line with previous studies that LRG1 have the role of promoting proliferation in keratinocyte [35], colorectal cancer cells [36], non-small cell lung cancer cells [31] and pancreatic ductal adenocarcinoma [17]. LRG1 promoting GC proliferation suggests its oncogenic role in gastric cancer.
It has been reported that high levels of LRG1 facilitate migration [37,38,39], metastasis [17, 18], epithelial–mesenchymal transition (EMT) [18, 35, 38] in some malignant tumors, and benign diseases. LRG1 has been predicted to bind extracellular matrix proteins due to its leucine-rich repeats and tendency [40]. LRG1 was reported to be a new regulator of angiogenesis that mediates its effect by modulating TGF-β signaling [13]. LRG1 has been identified as a potential novel pathogenic mediator of angiogenesis in diabetic glomerular disease [41] and progression of osteoarthritis [39]. Angiogenesis is a process by which cancers supply the tumor microenvironment with nutrients and oxygen, thereby contributing to tumor growth [42]. It is still unclear whether LRG1 can regulate angiogenesis in gastric cancer and become a promising anti-angiogenic therapeutic target. In this study, we first showed that LRG1 plays a facilitative role in regulating tumor angiogenesis of GC. LRG1 in the conditioned medium of GC cells can regulate endothelial cell proliferation, migration and tube formation, which are all key indicators of angiogenesis.
We identified transcription factor ATF3 as a positive regulator of LRG1. This is in agreement with previous evidence that endothelial proliferation requires activation of stress response gene ATF3, and deletion of ATF3 reduced endothelial proliferation and compromised the regeneration [26]. Previous study has demonstrated that ATF3 was inhibitied by miR-494 [43]. LRG1 has also been described as a miR-494 target [44] opening the possibility that miR-494 controls LRG1 both directly and indirectly, via ATF3 suppression. ATF3 upregulation in gastric cancer highlights the clinical relevance of the ATF3–LRG1 axis.
Our results found that LRG1 promoted the activation of VEGFA, which was the first described cytokine that promoted tumor angiogenesis [45]. VEGFA is the main survival factor of ECS, which stimulates proliferation and migration, inhibits apoptosis, and regulates its permeability [46]. Previous studies have shown that STAT3 can transcriptionally activate VEGFA and promote angiogenesis [47, 48]. The transcription factor STAT3 has previously been shown to be downstream of the SRC pathway [49, 50]. STAT3 plays an important role in the genesis and development of tumors by regulating the expression of several genes and promoting cell proliferation, invasion, immunosuppression and angiogenesis [51]. In this study, we revealed that LRG1 activates the downstream signaling pathway SRC/STAT3/VEGFA axis. SRC /STAT3/VEGFA was identified as a signaling axis that may augment LRG1-mediated promotion of gastric cancer angiogenesis. In the future, it would be interesting to identify whether they can bind directly to each other, and to explore whether LRG1/SRC /STAT3 causes other cancer features, such as migration and invasion, or avoids gastric cancer immune damage, which also need to be studied.
In summary, this is a preliminary study to investigate the role of LRG1 in the angiogenesis of gastric cancer. Our data suggest a novel mechanism whereby LRG1 promotes GC proliferation and angiogenesis. Thus, LRG1 may be a potential biomarker and therapeutic target for gastric cancer.
Conclusions
In conclusion, our data unraveled a novel mechanism whereby LRG1 promotes GC proliferation and angiogenesis. These findings indicate LRG1 to be a potential new diagnostic and prognostic marker in GC.
Availability of data and materials
Please contact the corresponding author for all data requests.
Abbreviations
- LRG1:
-
Leucine-rich-alpha-2-glycoprotein 1
- ATF3:
-
Activating transcription factor 3
- GC:
-
Gastric cancer
- HUVECs:
-
Human umbilical vein endothelial cells
- CAM:
-
Chick embryo chorioallantoic membrane
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
References
Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. 2020;396:635–48.
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Xu Z, Hu C, Yu J, et al. Efficacy of conversion surgery following apatinib plus paclitaxel/S1 for advanced gastric cancer with unresectable factors: a multicenter, single-arm, phase II trial. FrontPharmacol. 2021;12:642511.
Nienhuser H, Schmidt T. Angiogenesis and anti-angiogenic therapy in gastric cancer. Int J Mol Sci. 2017;19:43.
Yoshida K, Yamaguchi K, Okumura N, et al. Is conversion therapy possible in stage IV gastric cancer: the proposal of new biological categories of classification. Gastric Cancer. 2016;19:329–38.
Fukuchi M, Ishiguro T, Ogata K, et al. Prognostic role of conversion surgery for unresectable gastric cancer. Ann Surg Oncol. 2015;22:3618–24.
Macedo F, Ladeira K, Longatto-Filho A, et al. Gastric cancer and angiogenesis: is VEGF a useful biomarker to assess progression and remission? J Gastric Cancer. 2017;17:1–10.
Saeed A, Park R, Sun W. The integration of immune checkpoint inhibitors with VEGF targeted agents in advanced gastric and gastroesophageal adenocarcinoma: a review on the rationale and results of early phase trials. J Hematol Oncol. 2021;14:13.
Haupt H. Baudner S [Isolation and characterization of an unknown, leucine-rich 3.1-S-alpha2-glycoprotein from human serum (author’s transl)]. Hoppe-Seyler’s Z Physiol Chem. 1977;358:639–46.
Takahashi N, Takahashi Y, Putnam FW. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc Natl Acad Sci USA. 1985;82:1906–10.
Serada S, Fujimoto M, Terabe F, et al. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2169–79.
Lynch J, Fay J, Meehan M, et al. MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-beta signalling pathway. Carcinogenesis. 2012;33:976–85.
Wang X, Abraham S, McKenzie JAG, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499:306–11.
Zhong D, He G, Zhao S, et al. LRG1 modulates invasion and migration of glioma cell lines through TGF-beta signaling pathway. Acta Histochem. 2015;117:551–8.
Zhong ME, Chen Y, Xiao Y, et al. Serum extracellular vesicles contain SPARC and LRG1 as biomarkers of colon cancer and differ by tumour primary location. EBioMedicine. 2019;50:211–23.
Serada S, Fujimoto M, Ogata A, et al. iTRAQ-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Ann Rheum Dis. 2010;69:770–4.
Xie ZB, Zhang YF, Jin C, et al. LRG-1 promotes pancreatic cancer growth and metastasis via modulation of the EGFR/p38 signaling. J Exp Clin Cancer Res CR. 2019;38:75.
Zhang J, Zhu L, Fang J, et al. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res CR. 2016;35:29.
Li Y, Zhang Y, Qiu F, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32:1976–83.
Linden M, Lind SB, Mayrhofer C, et al. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 2012;12:135–44.
Andersen JD, Boylan KL, Jemmerson R, et al. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res. 2010;3:21.
Wen SY, Zhang LN, Yang XM, et al. LRG1 is an independent prognostic factor for endometrial carcinoma. Tumour Biol. 2014;35:7125–33.
Yamamoto M, Takahashi T, Serada S, et al. Overexpression of leucine-rich alpha2-glycoprotein-1 is a prognostic marker and enhances tumor migration in gastric cancer. Cancer Sci. 2017;108:2052–60.
He L, Wang W, Shi H, et al. THBS4/integrin alpha2 axis mediates BM-MSCs to promote angiogenesis in gastric cancer associated with chronic Helicobacter pylori infection. Aging. 2021;13:19375–96.
Uloza V, Kuzminiene A, Palubinskiene J, et al. Model of human recurrent respiratory papilloma on chicken embryo chorioallantoic membrane for tumor angiogenesis research. Histol Histopathol. 2017;32:699–710.
McDonald AI, Shirali AS, Aragon R, et al. Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell. 2018;23:210-225e6.
Xiao S, Zhu H. Leucine-rich alpha-2-glycoprotein1 gene interferes with regulation of apoptosis in leukemia KASUMI-1 Cells. Med Sci Monit. 2018;24:8348–56.
Hatta W, Gotoda T, Koike T, et al. History and future perspectives in Japanese guidelines for endoscopic resection of early gastric cancer. Dig Endosc. 2020;32:180–90.
Japanese Gastric Cancer A. Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer. 2014;2017(20):1–19.
Smyth EC. Chemotherapy for resectable microsatellite instability-high gastric cancer? Lancet Oncol. 2020;21:204.
Li Z, Zeng C, Nong Q, et al. Exosomal leucine-rich-alpha2-glycoprotein 1 derived from non-small-cell lung cancer cells promotes angiogenesis via TGF-beta signal pathway. Mol Ther Oncol. 2019;14:313–22.
Fahrmann JF, Schmidt CM, Mao X, et al. Lead-time trajectory of CA19-9 as an anchor marker for pancreatic cancer early detection. Gastroenterology. 2021;160:1373-1383e6.
Zhang N, Ren Y, Wang Y, et al. LRG1 suppresses migration and invasion of esophageal squamous cell carcinoma by modulating epithelial to mesenchymal transition. J Cancer. 2020;11:1486–94.
Zhang Y, Luo Q, Wang N, et al. LRG1 suppresses the migration and invasion of hepatocellular carcinoma cells. Med Oncol. 2015;32:146.
Liu C, Teo MHY, Pek SLT, et al. A multifunctional role of leucine-rich alpha-2-glycoprotein 1 in cutaneous wound healing under normal and diabetic conditions. Diabetes. 2020;69:2467–80.
Zhou Y, Zhang X, Zhang J, et al. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS ONE. 2017;12:e0175122.
Gao Y, Xie Z, Ho C, et al. LRG1 promotes keratinocyte migration and wound repair through regulation of HIF-1alpha stability. J Investig Dermatol. 2020;140:455-464e8.
Ban Z, He J, Tang Z, et al. LRG1 enhances the migration of thyroid carcinoma cells through promotion of the epithelialmesenchymal transition by activating MAPK/p38 signaling. Oncol Rep. 2019;41:3270–80.
Wang Y, Xu J, Zhang X, et al. TNF-alpha-induced LRG1 promotes angiogenesis and mesenchymal stem cell migration in the subchondral bone during osteoarthritis. Cell Death Dis. 2017;8:e2715.
Saito K, Tanaka T, Kanda H, et al. Gene expression profiling of mucosal address in cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J Immunol. 2002;168:1050–9.
Hong Q, Zhang L, Fu J, et al. LRG1 promotes diabetic kidney disease progression by enhancing TGF-beta-induced angiogenesis. J Am Soc Nephrol. 2019;30:546–62.
Oya Y, Hayakawa Y, Koike K. Tumor microenvironment in gastric cancers. Cancer Sci. 2020;111:2696–707.
Cao Y, Mi X, Zhang D, et al. Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury. Clin Sci. 2020;134:139–54.
Su Q, Lv XW, Sun YH, et al. MicroRNA-494 inhibits the LRG1 expression to induce proliferation and migration of VECs in rats following myocardial infarction. Mol Ther Nucl Acids. 2019;18:110–22.
Leung DW, Cachianes G, Kuang WJ, et al. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–9.
Korpanty G, Smyth E, Carney DN. Update on anti-angiogenic therapy in non-small cell lung cancer: are we making progress? J Thorac Dis. 2011;3:19–29.
Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8.
Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–109.
Tan Q, Wang H, Hu Y, et al. Src/STAT3-dependent heme oxygenase-1 induction mediates chemoresistance of breast cancer cells to doxorubicin by promoting autophagy. Cancer Sci. 2015;106:1023–32.
Aggarwal BB, Sethi G, Ahn KS, et al. Targeting signal-transducer-and-activator-of-transcription-3 for prevention and therapy of cancer: modern target but ancient solution. Ann N Y Acad Sci. 2006;1091:151–69.
Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos.81402357).
Author information
Authors and Affiliations
Contributions
M.Y., Q.-C.D. and E.-D.Z. designed and analyzed the experimental data. L.-N.H., A.-Q.F., H.G. and H.-H.H. performed the experiments. L.-N.H. and M.Y. prepared the figures.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interests.
Ethical considerations
All procedures followed were in accordance with the ethical standards of the Ethical Committee of Tongji Medical College, Huazhong University of Science & Technology and with the Helsinki Declaration of 1964 and later versions. All human samples were obtained with the patients’ written informed consent. All institutional and national guidelines for the care and use of laboratory animals were followed.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10120_2022_1279_MOESM3_ESM.pdf
Supporting Information 3: Supplementary Figure S1. Immunofluorescence staining of VWF and CD31 for cell surface antigens of HUVECs
10120_2022_1279_MOESM4_ESM.pdf
Supporting Information 4: Supplementary Figure S2. (a)qRT-PCR of ATF3 and LRG1 mRNA levels after silencing of ATF3 in SGC7901 cells. (b) qRT-PCR of ATF3 and LRG1 mRNA levels after silencing of ATF3 in MFC cells
Rights and permissions
About this article
Cite this article
He, L., Feng, A., Guo, H. et al. LRG1 mediated by ATF3 promotes growth and angiogenesis of gastric cancer by regulating the SRC/STAT3/VEGFA pathway. Gastric Cancer 25, 527–541 (2022). https://doi.org/10.1007/s10120-022-01279-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01279-9