Abstract
Background
Long non-coding RNAs (lncRNAs) have emerged as important roles in gastric cancer (GC). However, the role of the dysregulated lncRNAs in GC remained large unknown. We investigated the clinical significance, biological function and mechanism of CTC-497E21.4 in GC.
Methods
Firstly, RTFQ-PCR was used to detect the expression of CTC-497E21.4 in GC. Furthermore, knockdown of CTC-497E21.4 was conducted to assess the effect of CTC-497E21.4 in vitro and vivo. Subcellular localization of CTC-497E21.4 was determined by nuclear plasmolysis PCR and FISH. We also predicted CTC-497E21.4 binding miRNAs and downstream target genes and evaluated its regulation of miR-22 by acting as a ceRNA.
Result
CTC-497E21.4 was upregulated in GC tissues and GC cell lines (P < 0.05), and the expression was associated with depth of invasion, lymph node metastasis, and neurological invasion. Besides, knockdown of CTC-497E21.4 inhibited cell proliferation, invasion and promoted cell cycle arrest in vitro and inhibited tumorigenesis in vivo. Mechanistic investigations indicated that CTC-497E21.4 acted as a ceRNA for miR-22 and regulated NET1 expression. CTC-497E21.4/miR-22-3p/NET1 participated in the RhoA signaling pathway in the GC progression.
Conclusion
CTC-497E21.4 competed with miR-22 to regulate the expression of NET1 and regulated the malignant progression of GC through RhoA signaling pathway.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one of the most common cancers in the world. 2018 cancer statistics showed that GC remains an important cancer worldwide and is responsible for over 1,000,000 new cases and 783,000 dead cases. GC is the fifth most frequently diagnosed cancer and the third leading cause of cancer death [1, 2]. Although the incidence of GC has declined in recent years, GC remains a major cause of morbidity and mortality worldwide, especially in China [3, 4]. Due to the lack of sensitive and specific tumor markers, more than 80% of patients are diagnosed at an advanced stage with extensive and invasive lymph nodes and distant metastases, and the 5-year survival rate is low [5]. As a result, it is necessary to explore the mechanism of GC tumorigenesis to find effective diagnostic biomarkers.
In 2011, Salmena et al. proposed a competitive endogenous RNA (ceRNA) hypothesis that any RNA molecule containing an miRNA response element (MRE) can bind to microRNAs (miRNAs) via MRE and affect miRNA-induced gene silencing, adjusting its function. Studies of solid tumors and hematopoietic malignancies have shown that ceRNAs played important roles in cancer tumorigenesis by altering the key oncogenic or tumor suppressor genes [6]. Recently, long non-coding RNAs (lncRNAs), which are with over 200 nucleotides in length, have been reported to function as ceRNAs and were involved in tumorigenesis and cancer metastasis through silencing tumor suppressors or activation of oncogenes via different mechanisms [7, 8].The "lncRNA-miRNA-mRNA" network has been a potential therapeutic target in human cancers, including GC[9]. For example, silencing lncRNA NNT-AS1 suppressed GC tumorigenesis and induced cell cycle arrest at G0/G1 phase via sponging miR-204/E2F1 [10]. Xu et al. revealed that SP1 could induce lncRNA SNHG1 and knockdown of SNHG1 repressed cell growth. Mechanistically, lncRNA SNHG1 in the nucleus could directly interact with PRC2 and lncRNA SNHG1 could also act as sponge for miR-145-5p and suppress cyclin D2 expression. LncRNA SNHG1 can be a potential target for colorectal cancer [11].
Previously we screened novel lncRNAs via consulting literature [12] and analyzing GEO databases and the result indicated that CTC-497E21.4 was definitely upregulated in GC [13]. CTC-497E21.4 was also named linc00958 and highlighted to be an oncogene to promote bladder cancer progression [14, 15]. Our study showed that CTC-497E21.4 was highly expressed in the serum of GC patients. The expression levels of CTC-497E21.4 were associated with TNM stage and differentiation. High expression level of CTC-497E21.4 was related to poor outcomes [13]. The molecular mechanisms underlying the oncogenic functions of CTC-497E21.4 require further exploration.
MiR-22 was reported to be a tumor suppressor in various types of cancers including GC [16,17,18]. Studies showed that miR-22 inhibited GC cell proliferation, invasion and metastasis via upregulating MMP14 and Snail, and then inducing extracellular matrix (ECM) remodeling and EMT [19]. Neuroepithelial cell transforming gene 1 (NET1) is a guanine nucleotide exchange factor (GEF) which specifically activates RhoA protein and, thus, also involves in a range of biological processes including cell proliferation and apoptosis [20, 21]. Moreover, NET1 can also activate signaling pathways such as JNK, NF-κB and Wnt [22]. However, the mechanism of CTC-497E21.4 regulating miR-22/NET1 axis in GC remains unknown.
In our present study, CTC-497E21.4 acts as an oncogene in GC tumorigenesis to promote cell proliferation, invasion and metastasis. CTC-497E21.4 is mainly located in the cytoplasm and functions as a competing endogenous RNA (ceRNA) of miR-22 and targets NET1. CTC-497E21.4/miR-22/NET1 axis may be a potential novel therapeutic target for the treatment of GC.
Materials and methods
Clinical GC tissues
A total of 60 GC tissues and paired normal tissues were obtained from resection before receiving any other therapy at Affiliated Hospital of Nantong University (Nantong, China) between June 2017 and July 2018. The clinical characteristics of all patients were recorded and summarized in Supplementary Tables S1. This study was approved by the Clinical Research Ethics Committee of the Affiliated Hospital of Nantong University.
Cell lines and culture
All the human GC cell lines (SGC-7901, MKN-45, HGC-27 and AGS) and gastric epithelial cell line (GES-1) were obtained from the Chinese Academy of Sciences Committee on Type Culture Collection Cell Bank (Shanghai, China). GC cell lines were all cultured in 1640 medium (Corning, USA) and GES-1 was cultured in DMEM medium (Corning, USA) with 10% Fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin–streptomycin in a humid condition with 5% of CO2 at 37 °C.
RNA isolation and real-time fluorescent quantitative polymerase chain reaction (RTFQ-PCR)
Total RNA was extracted from GC tissues and cultured cell lines using Trizol Reagent (Invitrogen, CA, USA). The concentration and purity of RNA were determined by micro-spectrophotometer (Thermo Scientific, USA). 1 μg total RNA reversed transcription into complementary DNA (cDNA) using the Revert Aid First-Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA). RTFQ-PCR was performed by Roche LightCycler 480 (Roche, Switzerland) according to the manufacturer’s protocol using SYBR Green Master Mix (Roche, GER). 18S rRNA and U6 were used as internal control when detecting the expression of these genes. The primers of miR-22 and U6 were purchased from RiboBio (RiboBio, Guangzhou, China). The relative gene expression was calculated using the 2−ΔΔCt method. The specific primer sequences used in this study are presented in Supplementary Table S2.
Tissue fluorescence in situ hybridization (FISH)
The paraffin sections were pretreated before incubating with FISH probe in hybridization buffer at 37 °C overnight and the slide was washed and dehydrated at 42 °C for three times. Finally, DNA staining was performed and observed under a fluorescence microscope. RNA FISH probes were designed by RiboBio (RiboBio, Guangzhou, China).
Cell transfection
CTC-497E21.4 short hairpin RNAs (shRNAs) (sh-CTC-497E21.4), pcDNA-CTC-497E21.4 and miR-22 inhibitor were purchased from GenePharma (GenePharma, Suzhou, China). Short interference RNA (siRNA) for NET1 (si-NET1) was designed by RiboBio (RiboBio, Guangzhou, China). The transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. At 48 h or 72 h post-transfection, GC cells were harvested for further study. The shRNA, siRNA and pcDNA sequences used in the transfection are listed in Supplementary Table S3.
Cell proliferation, cell cycle analysis, migration and invasion
Cell Counting Kit-8 (CCK-8) assay and colony formation assay were conducted to perform the ability of cell proliferation. A CCK-8 assay kit (Dojindo Japan) was used to measure cell viability. 3000 transfected cells were grown in the 96-well plates and 10 μl CCK-8 solution was added to each well; the absorbance was measured at 450 nm every 24 h. All the experiments were repeated in five replicates. For the colony formation assay, a total number of 600 transfected cells were seeded in six-well plates and the cells were cultured for about 14 days. Cell colonies were washed with phosphate-buffered saline (PBS) and stained with 0.1% crystal violet. Colonies containing more than 50 cells were counted and the mean colony numbers were calculated.
Flow cytometric analysis was performed to analyze cell cycle distribution. Briefly, GC cell lines treated with different plasmids for 48 h. At the indicated time point, adherent cells were collected, washed twice with ice-cold PBS, 500 μl ice-cold 70% ethanol used to fix cells, incubated at 4 °C for the night, and treated cells using cell cycle kit (Solarbio, China). The DNA content of the stained nuclei was analyzed for the cell cycle phase by BD FACS Calibur flow cytometer (BD Biosciences, San Diego, CA, USA).
Transwell assay and wound healing assay were used to detect cell invasion and migration. 50,000 GC cells were seeded into the upper transwell chambers (Corning). For invasion assay, cells were added into the upper chamber precoated with matrigel (BD Bioscience). In both assays, cells were maintained in the medium without serum in the upper chamber, and medium containing 20% FBS was added to the lower chamber as chemoattractant. After 24 h incubation, the transwell chambers were fixed with 4% paraformaldehyde, washed with PBS, and stained with 0.1% crystal violet. Three random fields were counted per chamber using a microscope (Olympus). All the experiments repeated three times.
CTC-497E21.4 subcellular location analysis
GC cell lines were fixed in 4% paraformaldehyde for 30 min and followed by washes with PBS. Fixed cells were treated with triton X-100 (Solarbio, China) and subsequently treated with Fluorescent In Situ Hybridization Kit (RiboBio, China) following the manufacturers’ instructions. Cytoplasmic and nuclear RNA was separated and purified using the PARIS Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturers’ instructions. RTFQ-PCR was used to detect the relative expression.
Luciferase reporter assay
The full-length CTC-497E21.4 cDNA was cloned into pSI-Check2 vector (Promega). The pSI-Check2-CTC-497E21.4-wild vector or pSI-Check2- CTC-497E21.4-mut vector and miR-22 mimics were co-transfected into 293 T cell by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Similarly, pSI-Check2-NET1-wild or pSI-Check2-NET1-mut and miR-22 mimics were co-transfected into 293 T cell by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). After 48 h, the activity of Renilla plasmid (Promega) was measured using Dual-Luciferase Reporter Assay Kit (Promega).
Tumor xenografts in animals
The xenograft mice in vivo assays were done according to the institutional guidelines and approved by the Animal Ethics Committee of Affiliated Hospital of Nantong University. Four-week-old male null mice (about 18 g) were purchased from the animal center of Nantong University (Nantong, China). The null mice were divided into two groups, SGC-7901 cells (transfected with NC plasmid or sh-CTC-497E21.4, 1 × 107/ml, 200 μl) were injected subcutaneously. The volume of xenograft tumors was measured every 5 days. After 25 days, the neoplasm was excised for weighting. The xenograft tumors were cut and subjected to H&E and immunohistochemical staining.
Western blotting analysis
Proteins were isolated from SGC-7901 and MKN-45 cell lines as previously treated using Native lysis Buffer (Solarbio, Beijing Solarbio Science and Technology, Beijing China). Cell protein lysates were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Specific bands were detected by Immunoblots visualized by ECL detection system (Quantity One software, BioRad). GAPDH antibody (1:10,000) (Abcam, Cambridge, MA, USA) was used as a control. NET1 (1:500) and RhoA (1:500) antibodies were purchased from Proteintech (Proteintech, China), Rac1, CDC42 (1:1000) and Phospho-Rac1/CDC42 (Ser71) (1:500) were purchased from Cell Signaling Technology (Cell Signaling Technology, USA). Moreover, Phospho-RhoA (phospho S188) (1:1000) was obtained from Abcam.
Statistical analysis
All data were analyzed using SPSS 20.0 software (IBM SPSS Statistics, Chicago, USA). Independent-Samples t test was used to analyze relative expression of GC tissues, one-way analysis of variance (ANOVA) was used to analyze multiple groups, and a Chi-square test was used for clinicopathological features statistical analysis. P < 0.05 was determined statistical significance. The figures were drawn using Graphpad 7.0.
Results
CTC-497E21.4 was upregulated in GC and indicated poor survival
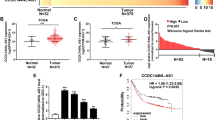
RTFQ-PCR was used to detect the expression of CTC-497E21.4 in 60 GC tissues and paired adjacent normal tissues, and the result showed that the expression of CTC-497E21.4 was significantly upregulated in 60 GC tissues compared with adjacent normal tissues (P = 0.0012, Fig. 1a). The cohort of GC patients was divided into high and low expression groups according to the mean value. We also analyzed the correlation between the CTC-497E21.4 expression and clinicopathological characteristics. The results indicated that dramatically high levels of CTC-497E21.4 were present in patients with advanced GC stage, lymph node metastasis and neural invasion (Table 1). What is more, survival analysis downloaded from KM plotter database whose data sources contain TCGA database and GEO databases indicated that CTC-497E21.4 overexpression was significantly correlated with poor overall survival (OS) (Fig. 1b). Moreover, four GC cell lines (SGC-7901, MKN-45, HGC-27, and AGS) showed significantly higher CTC-497E21.4 levels than the gastric epithelium cell line (GES-1) (Fig. 1c). In addition, we used FISH analysis to show CTC-497E21.4 expression in mice xenograft tissues and the result showed that the fluorescence expression of CTC-497E21.4 was stronger in NC group than that in interference group (Fig. 1d). These results indicated that CTC-497E21.4 upregulation may play a critical role in the development and progression of GC.
CTC-497E21.4 was significantly upregulated in GC tissues and GC cell lines. a Relative expression of CTC-497E21.4 in GC tissues and adjacent normal tissues. b Kaplan–Meier survival curve of patients with GC downloaded from TCGA database. c Relative expression of CTC-497E21.4 in GC cell lines and normal gastric epithelial cell line. d FISH analysis in the xenograft tumor tissues. *P < 0.05, **P < 0.01, ***P < 0.001
CTC-497E21.4 promoted cell proliferation and regulated cell cycle in GC
Because the expression of CTC-497E21.4 was relatively higher in SGC-7901 and MKN-45 cell lines, we transfected shRNA vector in these two cell lines and an overexpression plasmid was transfected in the AGS cell line (Figure S1). The results showed that sh-CTC-497E21.4 (2#, 3#) had higher knockdown efficiency, so we chose sh2 and sh3 for further study. CCK-8 assay and colony formation assay indicated that CTC-497E21.4 silencing inhibited the proliferation of SGC-7901 and MKN-45 cells (Fig. 2a–d), while CTC-497E21.4 overexpression promoted the proliferation of AGS cells (Fig. S2). Meanwhile, flow cytometry analysis demonstrated that compared with negative control (NC), CTC-497E21.4 silencing also showed cell cycle arrest in both SGC-7901 and MKN-45 cells lines. Significantly more cells were in G1/G0 phase and fewer cells were in S phase of the cell cycle (Fig. 2e, f), while enhanced CTC-497E21.4 expression produced the opposite effect (Fig. S3). Consistent with the cell cycle progression data, the expression levels of cell cycle proteins (such as Cyclin D1, p27) were markedly changed when CTC-497E21.4 was knocked down or enhanced (Figs. 2g, S3). These data indicated that CTC-497E21.4 could promote cell proliferation and silencing CTC-497E21.4 could be attributed to cell cycle arrest at the G1/G0 phase.
CTC-497E21.4 promoted GC cell proliferation and induced cell cycle arrest. a, b CCK-8 assay presented the proliferation of SGC-7901 and MKN-45 cells transfected with sh-CTC-497E21.4 or sh-NC. c, d Colony formation assay was performed to determine the proliferation of SGC-7901 and MKN-45 cells transfected with sh-CTC-497E21.4 or sh-NC. e, f Flow cytometry was used to determine the effect of sh-CTC-497E21.4 on SGC-7901 and MKN-45 cell cycle. g Western blot was used to detect the influence of CTC-497E21.4 silenced on cell cycle markers. *P < 0.05
CTC-497E21.4 contributed to the invasion and metastasis of GC cell lines
The transwell invasion assay analysis and wound healing assay found that knockdown of CTC-497E21.4 reduced the number of invaded SGC-7901 and MKN-45 cells and the migration speed was decreased (Fig. 3a–d). Conversely, enhancing CTC-497E21.4 produced the opposite effect (Figure S4). These findings inferred that CTC-497E21.4 was involved in cell invasion and migration in GC.
Knockdown of CTC-497E21.4 repressed SGC-7901 and MKN-45 cell invasion and migration. a, b Transwell assay was applied to measure the effect of knockdown CTC-497E21.4 deletion on cell invasion and migration capacity. c, d Wound healing assay was used to determine the motility of SGC-7901 and MKN-45 cells transfected with sh-CTC-497E21.4 and sh-NC. *P < 0.05
CTC-497E21.4 silencing inhibited tumor growth in vivo
To determine whether CTC-497E21.4 could affect the tumorigenesis of GC in vivo, SGC-7901 cells with NC or shRNA vector were subcutaneously injected into nude mice. The results showed that the weight of tumors in the interference group was smaller than that in the control group (Fig. 4a–c). The weight of xenograft tumors in the interference group was significantly lower than that in the negative control group (Fig. 4d). RTFQ-PCR analysis confirmed that CTC-497E21.4 expression of tumor tissues in the interference group was lower than that in negative control group (Fig. 4e). Furthermore, immunohistochemical (IHC) staining showed that the expression of the proliferation marker Ki-67 and transfer marker MMP9 was weaker in the group which was transfected with shRNA (Fig. 4f). All these results indicated that knockdown of CTC-497E21.4 could inhibit tumor growth in vivo.
Knockdown of CTC-497E21.4 inhibited tumor growth in nude mice xenograft models. a, b Knockdown of CTC-497E21.4 inhibited the growth of transplanted SGC-7901 cell in nude mice. c Growth curves of xenograft tumors after injection of mice with sh-CTC-497E21.4 or sh-NC. The tumor volumes were measured every 5 days after inoculation. d Tumor weights from two groups are represented. e RTFQ-PCR detected relative expression in two groups. f Ki67 and MMP9 protein levels in tumor tissues from sh-CTC-497E21.4 or negative control groups. *P < 0.05
CTC-497E21.4 regulated NET1 via acting as a ceRNA of miR-22
FISH and subcellular fractionation analysis were used to detect the distribution of CTC-497E21.4 and the results showed that CTC-497E21.4 was more abundant in the cytoplasm (Fig. 5a, b). To identify the potential miRNA targets of CTC-497E21.4, Starbase, and miRcode, DIANA Tools were used to predict 4 miRNAs (miR-185, miR-761, miR-22, miR-490) which may act as biological targets of CTC-497E21.4 (Fig. 5c). RTFQ-PCR was conducted to validate the binding of candidate miRNAs with CTC-497E21.4 and selected miR-22 for further study. We also detected the expression of CTC-497E21.4 in GC cells transfected with miR-22 inhibitor to determine the direct interaction between CTC-497E21.4 and miR-22 (Figs. 5d, S5). And luciferase reporter assay confirmed this strong relationship between miR-22 and CTC-497E21.4 (Fig. 5e). Similarly, we predicted target genes of miR-22 by TargetScan and NET1 was selected to be the target of miR-22 (Figure S6). Luciferase reporter assay also validated the relationship between miR-22 and NET1 (Fig. 5f).
NET1 was a target gene of miR-22 and was regulated by CTC-497E21.4. a FISH analysis of the location of CTC-497E21.4 in the cytoplasm and nuclear fractions of SGC7901 cells. b RTFQ-PCR was used to detect CTC-497E21.4 subcellular fractionation. c Bioinformatics predicted four miRNAs interacted with CTC-497E21.4. d RTFQ-PCR was used to determine the interaction between CTC-497E21.4 and miR-22. e, f The luciferase reporter assay conformed a strong relationship between miR-22 and CTC-497E21.4 as well as that between miR-22 and NET1. g, h RTFQ-PCR detected the relative expression of miR-22 and NET1 in the GC tissues. i Spearman–Pearson correlation of CTC-497E21.4 and NET1 expression and Spearman–Pearson correlation of miR-22 and NET1 expression in GC tissues. j RTFQ-PCR detected the expression of NET1 in GC cells transfected with sh-CTC-497E21.4 and miR-22 inhibitor. k Western blot analysis of NET1 in SGC-7901 and MKN-45cells with knockdown of CTC-497E21.4 and/or miR-22 inhibitor. L: IHC analysis of NET1 in mice xenograft models. *P < 0.05, **P < 0.01, ***P < 0.001
We also found that miR-22 expression was significantly lower in GC tissues, whereas NET1 was upregulated in GC tissues (Fig. 5g, h). Moreover, a negative correlation between CTC-497E21.4 and miR-22 expression was detected in GC tissues (r = − 0.5059, P = 0.0043, Fig. 5i), as well as a positive correlation between CTC-497E21.4 and NET1 (r = 0.5436, P = 0.0019, Fig. 5i). The RTFQ-PCR and western blotting showed that silencing CTC-497E21.4 reduced the expression of NET1 while miR-22 inhibitor enhanced the expression of NET1 (Fig. 5j, k). We also performed IHC assay to verify NET1 expression level in xenograft tumors and NET1 was decreased in the group transfected with sh-CTC-497E21.4 (Fig. 5l). All these data showed that CTC-497E21.4 targeted the miR-22/NET1 in the GC cells as ceRNA.
CTC-497E21.4/miR-22/NET1 axis promoted GC cell proliferation and migration in vitro
We performed rescue assays to validate whether miR-22 and NET1 were involved in the CTC-497E21.4 mediated inhibition of proliferation, cell cycle, migration and invasion. CCK-8 assay and colony formation assay confirmed that knock down CTC-497E21.4 or NET1 decreased cell proliferation ability. Besides, miR-22 inhibitor reversed the effect of sh-CTC-497E21.4 or si-NET1 on reducing cell proliferation (Fig. 6a, b). The transwell assay and flow cytometry analysis also revealed similar results that silencing CTC-497E21.4 or NET1 could suppress cell invasion and cell cycle arrest in G1 phase, while co-transfection of miR-22 inhibitor and sh-CTC-497E21.4 significantly increased cell invasion and cell cycle progression (Fig. 6c, d).
Effects of CTC-497E21.4 or NET1 silencing on the inhibition of GC cell growth and migration could be rescued by miR-22 inhibitor in vitro. a, b Effect of miR-22 inhibitor, si-NET1 and sh-CTC-497E21.4 on proliferation of GC cell lines. c Effect of miR-22 inhibitor, si-NET1 and shCTC-497E21.4 on cell cycle of GC cell lines. d Effect of miR-22 inhibitor, si-NET1 and sh-CTC-497E21.4 on migration of GC cell lines. *P < 0.05, **P < 0.01, ***P < 0.001
CTC-497E21.4 modulated GC progression through RhoA signaling pathway
Since NET1 is a RhoA-specific guanine exchange factor (GEF) and RhoA is one of the most extensively investigated members of the Rho GTPase family of proteins, the RhoA signaling pathway has been associated with cell proliferation as well as tumor invasion and metastasis [23, 24]. We tested whether expression of NET1 stimulated RhoA signaling pathway. We transfected sh-CTC-497E21.4, si-NET1 and miR-22 inhibitor vectors into SGC-7901 cell lines and detected the changes in RhoA relative genes. As presented in Fig. 7a, there was a change trend that knockdown of sh-CTC-497E21.4 or si-NET1 could affect NET1, RhoA, CDC42 and Rac1 in mRNA level. In addition, the result of western blot showed that sh-CTC-497E21.4, miR-22 inhibitor, and si-NET1 not only regulated the expression of RhoA, CDC42, and Rac1 at the total protein level, but also showed corresponding changes in phosphorylated proteins (Fig. 7b). Therefore, we speculated that NET1 may work as a RhoGEF and participated in the activation of RhoA signaling pathway which was mediated by CTC-497E21.4/miR-22/NET1 axis. Taken together, our data indicated that CTC-497E21.4 modulated GC cell proliferation, cell cycle, invasion and metastasis via RhoA pathway (Fig. 7c).
CTC-497E21.4 regulated RhoA signaling pathway. a RTFQ-PCR assay detected the mRNA expression of RhoA, CDC42 and Rac1 in SGC-7901 cell lines. b Western blot assay determined the total and active protein level of NET1, RhoA, CDC42 and Rac1 in SGC-7901 cell lines. c Summary of the mechanism of CTC-497E21.4 in GC cell lines
Discussion
As tumor molecular mechanism research gradually progressed, a growing number of genes have been confirmed to play an important role in all sorts of cancer. These human genes do not function alone, but form an interaction network through interactions to regulate the homeostasis of the internal environment. Furthermore, ceRNA is a novel network of interactions that lncRNAs act as a molecular sponge to moderate miRNAs expression [25,26,27]. Recently, the networks of “lncRNA-miRNA-mRNA” interactions were determined to participate in various diseases and may be used to act as biomarker predictions for developing novel therapeutic approaches in GC [28]. For example, Zhang et al. determined that lncRNA MT1JP overexpression inhibited GC cell proliferation, migration invasion, and promoted GC cell apoptosis. MT1JP/miR-92a-3p/FBXW7 axis regulated GC tumor occurrence and progression. MT1JP might be a potential target for GC therapy and diagnostic biomarker [29]. A novel lncRNA UFC1 was identified in GC, and UFC1 was closely associated with poor outcomes. The identification of UFC1/miR-498/Lin28b signaling axis could be important roles of lncRNAs in GC progression and provides new targets for GC diagnosis, prognosis and therapy [30].
In the present study, our results found that CTC-497E21.4 expression was upregulated in GC tissues and GC cell lines, and the higher expression of CTC-497E21.4 was associated with tumor depth, lymph node metastasis and neural invasion. Furthermore, high expression of CTC-497E21.4 was significantly associated with poor outcomes according to TCGA database. In in vitro and in vivo assays, we investigated that knockdown of CTC-497E21.4 suppressed GC cell proliferation, however, the difference in the number of cells may inevitably affect the migration and invasion of GC cells. This may explain why the cells on day 1 or 2 already showed slight reduction of numbers. Knockdown of CTC-497E21.4 also induced cell cycle arrest at G1/G0 phase. Moreover, CTC-497E21.4 silencing also retarded GC growth in vivo. Overexpression of CTC-497E21.4 showed the opposite effect. All these data indicated that CTC-497E21.4 was critical for gastric carcinogenesis and can be used as a potential indicator for the diagnosis and prognosis of GC.
In subsequent studies on the mechanism, we first determined that CTC-497E21.4 was mainly located in the cytoplasm. Evidence showed that ceRNA was a post-transcriptional regulation that occurred in the cytoplasm [31, 32]. As a result, we considered that CTC-497E21.4 may regulate the progression of GC through the ceRNA mechanism. Bioinformatic assay predicted that CTC-497E21.4 could contain potential miR-22, miR-185, miR-490 and miR-761 binding sites. Combined with RTFQ-PCR and luciferase reporter assays, CTC-497E21.4 could interact with miR-22 in GC. miR-22 was reported to be a tumor suppressor in GC. Zuo et al. found that the lower expression of miR-22 promoted GC invasion and metastasis by enhancing MMP14 and Snail and then inducing ECM remodeling and EMT [19]. Generally, lncRNAs exerted function by acting as ceRNA of miRNAs and regulated the expression of their targets. Using online predicting database, we determined that NET1 was a target of miR-22. Luciferase reporter assays also verified that miR-22 targeted NET1 mRNA at its 3′UTR. NET1, a guanine exchange factor (GEF), whose expression was enhanced in GC was reported to be associated with cell invasion by activating RhoA signaling pathway [23, 33, 34]. In the chronic myeloid leukemia (CML), miR-22 overexpression or NET1 silencing decreased cell proliferation and alteration in cell cycle and miR-22 could interact with NET1 [35]. In our study, we also determined that miR-22 was downregulated while NET1 was upregulated in GC, and the expression of miR-22 and NET1 was shown to be negatively corrected. What’s more, the influence of GC cell proliferation, migration and cell cycle caused by CTC-497E21.4 silencing or siNET1 was reversed by co-transfecting sh-CTC-497E21.4 and miR-22 inhibitor. Several studies indicated that NET1 could regulate RhoA activity [23, 33]. Rho GTPase family was involved in the regulation of invasion and metastasis of tumor cells. Among them, RhoA, Rac1 and CDC42 were the three most known regulators and it was reported that inhibition of RhoA, Rac1 and CDC42 could lead to cell cycle arrest at G1 phase [36, 37]. In our study, transfecting sh-CTC-497E21.4, miR-22 inhibitor or si-NET1 may have a certain effect on RhoA, CDC42 and Rac1 mRNA levels in SGC-7901 cell lines, although it had no statistic difference. Furthermore, western blot also showed that NET1 not only affected Rho family members total protein level but also showed corresponding changes in phosphorylated proteins. Therefore, we speculated that NET1 worked as a RhoGEF and participated in RhoA pathway mediated by CTC-497E21.4/miR-22/NET1 axis.
In conclusion, CTC-497E21.4 was overexpressed in GC and was associated with advanced GC stage, lymph node metastasis and neural invasion, which predicted poor prognosis. In addition, CTC-497E21.4 promoted NET1 expression by directly targeting miR-22 to promote GC progression. The present study provides a novel sight for GC tumorigenesis and confirms the potential value of CTC-497E21.4 for GC diagnosis and therapies.
References
Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Zong L, et al. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606.
Van Cutsem E, et al. Gastric cancer. Lancet. 2016;388(10060):2654–64.
Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8.
Sun M, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–310.
Zong W, et al. Long non-coding RNA-mediated regulation of signaling pathways in gastric cancer. Clin Chem Lab Med. 2018;56(11):1828–37.
Guo LL, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21(41):11680–7.
Chen B, et al. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J Cell Mol Med. 2018;22(10):4751–9.
Xu M, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141.
Gu J, et al. Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma. Oncotarget. 2017;8(30):49201–16.
Zong W, et al. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019;57(7):1063–72.
Seitz AK, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7(1):395.
He W, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128(2):861–75.
Zhang X, et al. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol Res. 2017;50(1):27.
Zhang H, et al. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015;356(2 Pt B):781–90.
Wang X, et al. MiR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun. 2014;445(1):175–9.
Zuo QF, et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6:e2000.
Carr HS, et al. Regulation of focal adhesion kinase activation, breast cancer cell motility, and amoeboid invasion by the RhoA guanine nucleotide exchange factor Net1. Mol Cell Biol. 2013;33(14):2773–866.
Chen C, Ji Q. Net1, a therapeutic potential target for cancer. Hum Pathol. 2018;71:168.
Zhang Y, et al. Short interfering RNA targeting Net1 reduces the angiogenesis and tumor growth of in vivo cervical squamous cell carcinoma through VEGF down-regulation. J Cell Sci. 2017;65:113–22.
Murray D, et al. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br J Cancer. 2008;99(8):1322–9.
Zuo Y, et al. Contributions of the RhoA guanine nucleotide exchange factor Net1 to polyoma middle T antigen-mediated mammary gland tumorigenesis and metastasis. Breast Cancer Res. 2018;20(1):41.
Chen X, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24(8):2002–144.
Lu G, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37(1):289.
Hu Y, et al. Roles of competing endogenous RNAs in gastric cancer. Brief Funct Genomics. 2016;15(3):266–73.
Zhang Y, et al. Identification of an lncRNAmiRNAmRNA interaction mechanism in breast cancer based on bioinformatic analysis. Mol Med Rep. 2017;16(4):5113–200.
Zhang G, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87.
Zhang X, et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. 2018;37(1):134.
Cao Z, et al. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–94.
Bai Y, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–18848.
Dubash AD, et al. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS ONE. 2011;6(2):e17380.
Carr HS, et al. Interaction of the RhoA exchange factor Net1 with discs large homolog 1 protects it from proteasome-mediated degradation and potentiates Net1 activity. J Biol Chem. 2009;284(36):24269–80.
Ahmad HM, et al. miR-22 regulates expression of oncogenic neuro-epithelial transforming gene 1, NET1. Febs J. 2014;281(17):3904–19.
Wang W, et al. Anti-proliferative effect of auriculataoside A on B16 melanoma 4A5 cells by suppression of Cdc42-Rac1-RhoA signaling protein levels. J Nat Med. 2019;73(3):450–5.
Aspenstrom P. Activated Rho GTPases in cancer-the beginning of a new paradigm. Int J Mol Sci. 2018;19(12):3949.
Acknowledgments
This study was supported by the National Nature Science Foundation (Grant numbers 81201349, 81000775); Young medical key talents in Jiangsu province (Grant numbers QNRC 2016686, 2016687); Frontier and key technical innovation projects of Nantong (Grant number MS22015049); Nantong Science and Technology Plan Project (MS12017008-3, MS12018031). We are especially grateful to Rongrong Jing for the work her paid during the revision of the article.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study protocol was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Nantong University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zong, W., Feng, W., Jiang, Y. et al. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer 23, 228–240 (2020). https://doi.org/10.1007/s10120-019-00998-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-00998-w