Abstract
Background
Emerging evidence have illustrated the vital role of long noncoding RNAs (lncRNAs) long intergenic non-protein coding RNA 00511 (LINC00511) on the human cancer progression and tumorigenesis. However, the role of LINC00511 in breast cancer tumourigenesis is still unknown. This research puts emphasis on the function of LINC00511 on the breast cancer tumourigenesis and stemness, and investigates the in-depth mechanism.
Methods
The lncRNA and RNA expression were measured using RT-PCR. Protein levels were measured using western blotting analysis. CCK-8, colony formation assays and transwell assay were performed to evaluate the cell proliferation ability and invasion. Sphere-formation assay was also performed for the stemness. Bioinformatic analysis, chromatin immunoprecipitation (ChIP) and luciferase reporter assays were carried to confirm the molecular binding.
Results
LINC00511 was measured to be highly expressed in the breast cancer specimens and the high-expression was correlated with the poor prognosis. Functionally, the gain and loss-of-functional experiments revealed that LINC00511 promoted the proliferation, sphere-formation ability, stem factors (Oct4, Nanog, SOX2) expression and tumor growth in breast cancer cells. Mechanically, LINC00511 functioned as competing endogenous RNA (ceRNA) for miR-185-3p to positively recover E2F1 protein. Furthermore, transcription factor E2F1 bind with the promoter region of Nanog gene to promote it transcription.
Conclusion
In conclusion, our data concludes that LINC00511/miR-185-3p/E2F1/Nanog axis facilitates the breast cancer stemness and tumorigenesis, providing a vital insight for them.
Similar content being viewed by others
Introduction
Breast cancer is one of the most common cancers in women worldwide and is the leading cause of cancer-related death in women [1,2,3]. The primary method for treating breast cancer is surgery, chemotherapy, and/or radiation therapy, greatly improving the therapeutic effects [4]. Research has shown that stem cell characteristics play a role in the recurrence and regeneration of breast cancer [5]. A special subgroup of tumor cells exists in the tumor, called cancer stem cells (CSCs) or tumor-initiating cells. CSCs have the potential of self-renewal, multidirectional differentiation, infinite proliferation and tumor reconstruction [6]. They are regarded as the source of tumor development, differentiation, invasion and metastasis, radiochemotherapy resistance and recurrence [7]. Breast cancer stem cells (BCSCs) have been verified to be the vital promoting factor for the breast cancer cells proliferation and self-renewal ability [8].
Long noncoding RNAs (lncRNAs) play a role in epigenetic regulation in human pathophysiology [9,10,11] and a large have been found to participate in cancer tumorigenesis [12]. For example, FEZF1-AS1 modulates Nanog expression through sponging miR-30a, suggesting the regulation of FEZF1-AS1/miR-30a/Nanog [13]. Further, H19 acts as a competitive endogenous RNA for miRNA let-7 resulting in the release of hypoxia-inducible factor 1α (HIF-1α), leading to increased PDK1 expression, thereby regulating the cancer stem-like characteristics [14].
Long intergenic noncoding RNA 00511 (LINC00511) is an oncogene that influences tumor size, metastasis, and poor prognosis. LINC00511 binds histone methyltransferase EZH2 and specifies the histone modification pattern on p57 [15]. It also acts as an oncogene in squamous cell carcinoma and pancreatic ductal adenocarcinoma [16, 17]. In the pre-experiments, we have performed large scale screening for the differentially expressed lncRNAs for breast cancer. After filtration and consultation, we select the LINC00511 as the target.
In the present study, LINC00511 acts as an oncogenic RNA in breast cancer tumorigenesis. LINC00511 is located in the cytoplasm and functions as a miR-185-3p ‘sponge’ and targets E2F1. The transcription factor E2F1 binds with the promoter of Nanog to activate its expression at the transcriptional level. The LINC00511/miR-185-3p/E2F1/Nanog axis may have therapeutic potential for breast cancer stemness and tumorigenesis.
Materials and methods
Clinical subjects and specimens
A total of 39 cases of breast cancer subject specimens were recruited at Youjiang Medical College Affiliated Hospital. The tumor tissue was excised during surgery and a pathological classification was made and graded by two independent experienced pathologists. All participators provided informed consent and the procedures were approved by the Ethics Committee of the Youjiang Medical College Affiliated Hospital.
Cell lines and culture
All the cell lines were provided by the American Type Culture Collection (ATCC), including normal human breast epithelial cell (MCF-10A) and breast cancer cells (MDA-MB-468, MDA-MB-231, MDA-MB-453, MCF-7). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) and supplemented with 10% fetal bovine serum (FBS) in atmosphere containing 5% CO2 at 37 °C.
Cell transfection
The miR-185-3p inhibitor, E2F1 shRNA, and negative control (NC) were purchased from Gene-Pharma Cells Company. The LINC00511 and E2F1 sequence were sub-cloned into the pcDNA3.1 vector (Invitrogen) to enhance LINC00511 expression (pcDNA-LINC00511) and empty pcDNA3.1 vector (pcDNA-NC). The transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif, USA) according to the manufacturer’s instructions. Short hairpin RNA targeting LINC00511 (shRNA-LINC00511) and control shRNA (shRNA-NC) were provided by Santa Cruz Biotechnology (Santa Cruz, CA, USA). The shRNA sequences used in the transfection are presented in Additional file 1: Table S1.
RNA isolation and quantitative polymerase chain reaction
Total RNA was isolated from breast cancer tissue and cells using Direct-zol™ RNA kits (Zymo research, USA) following the manufacturer’s protocol. RNA concentration was checked using a NanoDrop 2000 spectrophotometer (Termo scientifc, USA). cDNA was produced and the reverse transcription (RT) reaction was done using PrimeScrip-RT Reagent Kit (Takara) and ABI7900 system (Applied Biosystems, USA). The relative gene expression (fold change) was calculated using the 2-ΔΔCt method. The primer sequences used in the PCR are presented in Additional file 2: Table S2.
Sphere-formation assay
Breast cancer cells MDA-MB-231, and MCF-7 that transfected with shRNAs or plasmids were seeded in the six-well culture plates (Corning, NY, USA). Cells (2 × 105) and were cultured in the serum-free DMEM medium with EGF, hFGF (Peprotech, USA), insulin, and penicillin/streptomycin (Gibco) added as previously described [18]. Spheroids clones were fixed and stained with crystal violet, and counted under light stereomicroscope (Olympus, Tokyo, Japan).
Proliferation CCK-8 and colony formation assay
CCK-8 assay was conducted using a CCK-8 assay kit (Dojindo Japan). The transfected cells were seeded into culture plates and the incubated cells were treated with 10 μl of CCK-8 reagent. Absorbance was measured at 450 nm.
Transwell invasion assays
The breast cancer cells were seeded on the member pre-coated with matrigel (BD Biosciences, San Jose, CA, USA) using 24-well transwell chamber (Corning). After 24 h of incubation, the cells on the upper surfaces were scraped, and the invaded cells were fixed with 4% paraformaldehyde and stained with Giemsa. Cells were then counted under a light microscope.
Western blot assay
Western blot was conducted as previously described [19]. The proteins were extracted from the breast cancer specimens using RIPA lysis buffer (Beyotime, Shanghai, China). BCA Protein Assay Kit was used to evaluate the protein concentration. The protein was separated from the sample buffer using SDS-PAGE, transferred into PVDF membranes and blocked with 5% skim milk for 1 h. The primary antibodies were provided by Abcam Company, including anti-Nanog (ab109250, 1:1000), anti-Oct4 (ab184665, 1:1000), anti-SOX2 (ab137385, 1:1000). The member was incubated at 4 °C overnight and then incubated with horseradish peroxidase-conjugated secondary antibodies. Immunoblots were visualized by ECL detection system (Pierce, Rockford, IL, USA).
Luciferase gene reporter assay
Luciferase reporter vectors containing the wild-type or mutant sequences towards the E2F1 binding of Nanog promoter region were constructed. The vectors were co-transfected with E2F1 into MDA-MB-231 cells by the Lipofectamine 2000 reagent (Thermo Fisher, USA). The activity of Renilla plasmid (Promega) was measured using Dual-Luciferase Reporter Assay Kit (Promega).
Chromatin immunoprecipitation (ChIP)
The ChIP assay was done using the EZ-ChIP™ Chromatin immunoprecipitation kit (Millipore, USA). 1 × 107 cells were incubated using anti-E2F1 (5 μg) antibody and complexes were digested. Finally, the purified DNA was measured using qRT-PCR for each Nanog promoter region site.
Xenograft in vivo analysis
The xenograft mice in vivo assays were done according to the institutional guidelines and approved by the Animal Ethics Committee of Youjiang Medical College Affiliated Hospital. Four-week-old male null mice (about 20 g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd., (Shanghai, China). Breast cancer cells that transfected with lentivirus (sh-NC or sh-LINC00511, 5 × 107 cells/ml, 0.1 ml) were injected subcutaneously. The tumor dimensions were measured every 3 days and the volume was calculated using formula π/6 (length×width2). After 3 weeks, mice were sacrificed by cervical dislocation and the neoplasm was excised for weighting.
Statistical analysis
Data are presented as mean ± SD and all experiments were performed in triplicate. The difference within groups was assessed using one-way analysis of variance and independent samples t-tests. Analysis was performed using SPSS software and graphed using GraphPad Prism software. P value less than 0.05 was considered significant.
Results
LncRNA LINC00511 is ectopically over-expressed in the breast cancer tissues and cells compared to normal tissue and cells
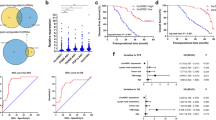
After comparison and screening, LINC00511 was found to be significantly up-regulated in the breast cancer tissues (Table 1). In the clinical samples, LINC00511 was over-expressed in the breast cancer tissue compared to normal adjacent tissue (Fig. 1a). There was a positive correlation between LINC00511 expression and clinicopathological parameters (TNM stages), with increased expression at more advanced stage of disease\ (Fig. 1b). The cohort of breast cancer patients was divided into high and low expression groups according to the mean value or the cutoff (Fig. 1c) and Kaplan-Meier analysis and log rank analysis were used to evaluate survival rate. Data illustrated that the patients with higher LINC00511 expression had poor prognosis compared to those with lower LINC00511 expression (Fig. 1d). These results suggest that lncRNA LINC00511 is ectopically over-expressed in breast cancer tissue and cells compared to normal tissue and cells, which means LINC00511 may be an oncogene for breast cancer.
LncRNA LINC00511 is ectopically over-expressed in breast cancer tissue compared to normal tissue. a LINC00511 expression in the breast cancer tissue compared with normal adjacent tissue. b LINC00511 expression in the breast cancer tissue with TNM stage. c High and low expression groups according to the mean value. d Survival rate of patients with high or low LINC00511 expression. *** indicates p value less than 0.001. * indicates p value less than 0.05
LncRNA LINC00511 promotes malignant cell proliferation and invasion in vitro and vivo
Colony formation assay and CCK-8 assay revealed that LINC00511 silencing inhibited the proliferation of MDA-MB-231 cells, while enhanced LINC00511 expression promoted the proliferation of MDA-MB-231 cells (Fig. 2a, b, c, d). The transwell invasion assay analysis found that LINC00511 silencing reduced the number of invaded MDA-MB-231 cells, while enhanced LINC00511 expression produced the opposite effect (Fig. 2e, f). Finally, xenograft model in mice assay showed that LINC00511 silencing inhibited the tumor growth (volume and weight) in vivo (Fig. 2g, h). Overall, results found that lncRNA LINC00511 promotes the proliferation and invasion of malignant cells both in vitro and vivo.
LncRNA LINC00511 promotes cell proliferation and invasion in vitro and vivo. a, b Colony formation assay in MDA-MB-231 and MCF-7 cells transfected with sh-LINC00511 or sh-NC. c, d CCK-8 assay presented the proliferation of MDA-MB-231 and MCF-7 cells transfected with sh-LINC00511 or sh-NC. e, f Number of invaded MDA-MB-2 31 cells and MCF-7 cells. g, h Tumor growth (volume and weight) in vivo. ** indicates p value less than 0.01. * indicates p value less than 0.05
LncRNA LINC00511 contributes to the maintenance of breast cancer CSC characteristic
RT-qPCR found that LINC00511 expression was over-expressed in the breast cancer cell lines (MDA-MB-468, MDA-MB-231, MDA-MB-453, MCF-7) compared to control lines (MCF-10A) (Fig. 3a). In the functional cellular experiments, shRNA targeting LINC00511 and enhanced expression plasmids were transfected into breast cancer cells MDA-MB-231, MCF-7 to knockdown or enhance expression (Fig. 3b, c). Western blot analysis revealed that LINC00511 silencing suppressed stem factor (Oct4, Nanog, SOX2) expression in MDA-MB-231 cells (Fig. 3d), while enhanced LINC00511 expression significantly increased stem factor expression in MCF-7 cells (Fig. 3e). Sphere-formation assay showed that LINC00511 silencing reduced mammosphere diameter and quantity (Fig. 3f, g), while enhanced LINC00511 expression significantly increased mammosphere diameter and quantity (Fig. 3h, i). Results show that LINC00511 contributes to the maintenance of breast cancer CSC characteristics, indicating the role of LINC00511 in breast cancer cell stemness.
LncRNA LINC00511 contributes to the maintenance of breast cancer CSC characteristic. a LINC00511 expression in breast cancer cell lines MDA-MB-468, MDA-MB-231, MDA-MB-453, and MCF-7 and normal cell lines (MCF-10A). b, c Breast cancer cell MDA-MB-231, and MCF-7 LINC00511 expression. d Stem factor (Oct4, Nanog, SOX2) expression in MDA-MB-231 cells transfected with shRNA for LINC00511. e The stem factors (Oct4, Nanog, SOX2) expression in MCF-7 cells transfected with enhanced LINC00511 plasmids. f, g Mammosphere diameter and quantity when transfected with shRNA for LINC00511. h, i Mammosphere diameter and quantity when transfected with enhanced LINC00511 plasmids. ** indicates p value less than 0.01. * indicates p value less than 0.05
LINC00511 targeted the miR-185-3p/E2F1 in the breast cancer cells as competing endogenous RNA
Mechanical analysis revealed that LINC00511 is positioned on the cytoplasm rather than the nulceus in breast cancer cells MDA-MB-231, MCF-7 (Fig. 4a). This finding suggests that LINC00511 exerts its downstream effects by post-transcriptional epigenetic regulation. Results confirmed that LINC00511 had several potential binding sites with the downstream miRNAs (miR-185-3p), and the luciferase reporter assays found a strong relationship between miR-185-3p and LINC00511 (Fig. 4b). The expression of miR-185-3p was decreased in breast cancer cells MDA-MB-231, and MCF-7 (Fig. 4c). Moreover, miR-185-3p was over-expressed in the LINC00511 silenced transfection, while decreased in the enhanced LINC00511 plasmid transfection (Fig. 4d). Results also confirmed that E2F1 acted as the target of miR-185-3p, acting as a functional protein of miR-185-3p, which was further validated by luciferase reporter assays (Fig. 4e). Similarly, we found that E2F1 mRNA expression was increased in breast cancer cells MDA-MB-231, and MCF-7 (Fig. 4f) and up-regulated in the miR-185-3p inhibitor transfection and enhanced LINC00511 plasmid transfection (Fig. 4g). These results suggest that LINC00511 targets the miR-185-3p/E2F1 in breast cancer cells as ceRNA.
LINC00511 targeted the miR-185-3p/E2F1 in the breast cancer cells as competing endogenous RNA. a The subcellular position of LINC00511 on the cytoplasm or nucleus. GAPDH and U1 acted as the cytoplasm and nuclear control. b The binding sites of miR-185-3p and LINC00511. c miR-185-3p expression in the breast cancer cells MDA-MB-231, and MCF-7 measured by RT-PCR. d miR-185-3p expression in the transfection of LINC00511 silencing and enhanced LINC00511 plasmid. e miR-185-3p and E2F1 3’-UTR binding sites. f E2F1 mRNA expression in the breast cancer cells MDA-MB-231, and MCF-7 measured by RT-PCR. g E2F1 mRNA expression in the miR-185-3p inhibitor (miR-185-3p inhib) transfection and enhanced LINC00511 plasmid transfection. ** indicates p value less than 0.01. * indicates p value less than 0.05
E2F1 enhanced the Nanog expression at the transcriptional level
The promoter region of Nanog gene was divided into four sections and shown in Fig. 5a. Chromatin immunoprecipitation (ChIP) assay revealed that E2F1 endogenous binds with the (Additional file 3) Nanog promoter region sites (− 1000~ − 400) (Fig. 5b). Then, we performed bioinformatics tools to predict the binding sites of E2F1 for Nanog gene promoter. The bioinformatics tools are as following, UCSC (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/), PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/), JASPAR (http://jaspar.genereg.net/). In the promoter region of Nanog gene (+ 100~ − 2000 nt), we found that transcription factor E2F1 could bind with the region (− 586~ − 576) using PROMO and JASPAR. The wild type or mutant sequences of Nanog promoter region (− 586 ~ − 576) were established (Fig. 5c). Results indicated that the luciferase activity of wild type sequences of Nanog promoter regions was increased, while the mutant type was not. This suggests the binding site is within E2F1 and Nanog promoter region (Fig. 5d). Western blot indicted that enhanced E2F1 plasmid transfection increased Nanog protein expression (Fig. 5e), while the E2F1 silencing decreased Nanog protein expression (Fig. 5f). Results show that E2F1 enhanced Nanog expression at the transcriptional level, providing a potential mechanism by which LINC0051/miR-185-3p/E2F1 axis promotes breast cancer stemness and tumorigenesis (Fig. 6).
E2F1 enhanced Nanog expression. a The Nanog gene promoter region. b Chromatin immunoprecipitation (ChIP) assay was performed to identify which region functioned as the effective binding site of Nanog promoter region. c The luciferase reporter plasmid having wild type or mutant sequences of Nanog promoter region (− 586 ~ − 576) were established. d The luciferase activity of wild type sequences and mutant of Nanog promoter region. e, f Nanog protein expression when transfected with enhanced E2F1 plasmid or E2F1 silencing
Discussion
Breast cancer is a one of the most common cancers in women worldwide [20, 21]. Although treatment for breast cancer has improved, it still represents a high disease morbisity and mortality. Epigenetic modification is as a significant regulatory mechanism for human pathogenesis [22] and long noncoding RNAs (lncRNAs) have been found to play a vital role.
In the tumourigenesis of breast cancer, the stem properties and self-renewal capability can trigger the recurrence and metastasis of breast cancer cells [23,24,25]. The role of lncRNAs on cancer cell stemness have been widely reported in human cancers. For example, SOX2OT is up-regulated in glioma tissue and glioblastoma stem cells, and knockdown of SOX2OT inhibits the proliferation, migration and invasion of cells, and the SOX2OT-miR-194-5p/miR-122-SOX3-TDGF-1 pathway forms a positive feedback loop for glioma cell stemness [26]. High levels of OIP5-AS1 can post-transcriptionally modulate downstream target genes with promoter binding motifs and are activated by stemness-associated transcription factors in cancer [27].
In the present study, results found that lncRNA LINC00511 expression was increased in breast cancer tissue samples and cell lines and may be related to poor prognosis. These results suggest that LINC00511 may be an oncogene for breast cancer. The role of LINC00511 on the functional phenotype and stemness of breast cancer cells was investigated. Results of gain and loss-of-functional assays revealed that LINC00511 promotes sphere-formation, stem factor (Oct4, Nanog, SOX2) expression, and contributes to the maintenance of breast cancer CSC characteristic, indicating the role of LINC00511 in breast cancer cell stemness. Finally, results found that LINC00511 promotes cell proliferation and invasion in vitro and vivo [28, 29].
Up to now, CSCs have been isolated from multiple tumors, such as breast cancer, lung adenocarcinoma, colorectal cancer, malignant melanoma, and glioma [30]. The invasion and metastasis of tumor is an important malignant biological phenotype of tumor. Recently, increasing studies have suggested that CSCs play a vital role in tumor invasion and metastasis [31].
Further experiments investigated the mechanism by which LINC00511 regulates breast cancer pathophysiology. Results found that LINC00511 functions as a miR-185-3p ‘sponge’ to harbor its expression, and then target E2F1 protein, confirming the role of LINC00511/miR-185-3p/E2F1 axis in breast cancer. Similar results have found that lncRNA H19 correlates positively with LIN28 by acting as a ceRNA for miRNA let-7, to form a double-negative feedback loop, thereby regulating the maintenance of breast cancer CSCs [32].
Finally, results show that E2F1 acts as a transcription factor that binds with the Nanog promoter region. Luciferase reporter assay, ChIP and western blot confirmed the direct action within E2F1 and Nanog, enriching the regulatory pathway to be LINC00511/miR-185-3p/E2F1/Nanog. The transcription factor E2F1 has been identified to be a oncogenic element in bladder cancer and colorectal cancer [33, 34]. E2F1 was also reported to up-regulated and involved in the carcinogenesis of breast cancer and results found it is involved in the expression of Nanog in breast cancer.
As regarding to the role of lncRNAs on the breast cancer CSCs properties, we discover the vital pathway of LINC00511/miR-185-3p/E2F1/Nanog. Moreover, emerging evidence and published papers also reveal the relevant findings. For example, lncRNA HOTAIR tightly regulates the proliferation, colony formation, migration and self-renewal capacity of breast cancer CSCs, and specifically inhibits miR-34a, which leads to the upregulation of Sox2 protein [35]. Thus, it is clear that lncRNAs could powerfully regulate the breast cancer stemness and tumorigenesis [36].
Conclusion
Results from this study show that LINC00511 acts as an oncogenic RNA in breast cancer tumorigenesis when present at high levels. LINC00511 functions as a miR-185-3p ‘sponge’ to harbor its expression, and then target E2F1 protein, which binds with the Nanog promoter region to activate its transcription. The characterization of the LINC00511/miR-185-3p/E2F1/Nanog axis provides an important insight for breast cancer stemness and tumorigenesis.
Abbreviations
- ceRNA:
-
Competing endogenous RNA
- ChIP:
-
Chromatin immunoprecipitation
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- lncRNA:
-
Long non-coding RNA
- PCR:
-
Polymerase chain reaction
- RIP:
-
RNA immunoprecipitation
References
Roumeliotis GA, Dostaler G, Boyd KU. Complementary and Alternative medicines and patients with breast Cancer: a case of mortality and systematic review of patterns of use in patients with breast Cancer. Plast Surg (Oakv). 2017;25:275–83.
Williams AD, Payne KK, Posey AD Jr, Hill C, Conejo-Garcia J, June CH, Tchou J. Immunotherapy for breast Cancer: current and future strategies. Curr Surg Rep. 2017;5:899–905.
Cagel M, Bernabeu E, Gonzalez L, Lagomarsino E, Zubillaga M, Moretton MA, Chiappetta DA. Mixed micelles for encapsulation of doxorubicin with enhanced in vitro cytotoxicity on breast and ovarian cancer cell lines versus Doxil((R)). Biomed Pharmacother. 2017;95:894–903.
Fan S, Yang Z, Ke Z, Huang K, Liu N, Fang X, Wang K. Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed Pharmacother. 2017;95:1636–43.
Shafei A, El-Bakly W, Sobhy A, Wagdy O, Reda A, Aboelenin O, Marzouk A, El Habak K, Mostafa R, Ali MA, Ellithy M. A review on the efficacy and toxicity of different doxorubicin nanoparticles for targeted therapy in metastatic breast cancer. Biomed Pharmacother. 2017;95:1209–18.
Zhou Y, Xia L, Wang H, Oyang L, Su M, Liu Q, Lin J, Tan S, Tian Y, Liao Q, Cao D. Cancer stem cells in progression of colorectal cancer. Oncotarget. 2018;9:33403–15.
Matteucci C, Balestrieri E, Argaw-Denboba A, Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018. https://doi.org/10.1016/j.semcancer.
Ding J, Wang X, Zhang Y, Sang X, Yi J, Liu C, Liu Z, Wang M, Zhang N, Xue Y, Shen L, Zhao W, Luo F, Liu P, Cheng H. Inhibition of BTF3 sensitizes luminal breast cancer cells to PI3Kalpha inhibition through the transcriptional regulation of ERalpha. Cancer Lett. 2018;440-441:54–63.
Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–8.
Tang T, Cheng Y, She Q, Jiang Y, Chen Y, Yang W, Li Y. Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed Pharmacother. 2018;107:338–46.
Wang Y, Hou Z, Li D. Long noncoding RNA UCA1 promotes anaplastic thyroid cancer cell proliferation via miR135amediated cmyc activation. Mol Med Rep. 2018;18:3068–76.
Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2018;00:1–9. https://doi.org/10.1111/jcmm.13987.
Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell Physiol. 2018;233:7234–45.
Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, Cui B, Wang HF, Zhao Y, An F, Guo T, Liu XF, Zhang L, Lv L, Lv DK, Xu LZ, Xie JJ, Lin WX, Lam EW, Xu J, Liu Q. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–74.
Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung Cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385.
Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47:468–76.
Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, Lin Q, Fu Z, Rufu C. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655–67.
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55.
Wang Y, Zhang L, Zheng X, Zhong W, Tian X, Yin B, Tian K, Zhang W. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382:137–46.
Xia E, Shen Y, Bhandari A, Zhou X, Wang Y, Yang F, Wang O. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res. 2018;8:1273–87.
Deng W, Wang Y, Zhao S, Zhang Y, Chen Y, Zhao X, Liu L, Sun S, Zhang L, Ye B, Du J. MICAL1 facilitates breast cancer cell proliferation via ROS-sensitive ERK/cyclin D pathway. J Cell Mol Med. 2018;22:3108–18.
Lin YF, Tseng IJ, Kuo CJ, Lin HY, Chiu IJ, Chiu HW. High-level expression of ARID1A predicts a favourable outcome in triple-negative breast cancer patients receiving paclitaxel-based chemotherapy. J Cell Mol Med. 2018;22:2458–68.
Lin X, Chen W, Wei F, Zhou BP, Hung MC, Xie X. Nanoparticle delivery of miR-34a eradicates long-term-cultured breast Cancer stem cells via targeting C22ORF28 directly. Theranostics. 2017;7:4805–24.
Park YH. The nuclear factor-kappa B pathway and response to treatment in breast cancer. Pharmacogenomics. 2017;18:1697–709.
Kubatka P, Uramova S, Kello M, Kajo K, Kruzliak P, Mojzis J, Vybohova D, Adamkov M, Jasek K, Lasabova Z, Zubor P, Fialova S, Dokupilova S, Solar P, Pec M, Adamicova K, Danko J, Adamek M, Busselberg D. Antineoplastic effects of clove buds (Syzygium aromaticum L.) in the model of breast carcinoma. J Cell Mol Med. 2017;21:2837–51.
Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J, Chen J, Liu L, Cai H, Li Z, Zhao L, He Q, Xue Y. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer. 2017;16:171.
Arunkumar G, Anand S, Raksha P, Dhamodharan S, Prasanna Srinivasa Rao H, Subbiah S, Murugan AK, Munirajan AK. LncRNA OIP5-AS1 is overexpressed in undifferentiated oral tumors and integrated analysis identifies AS a downstream effector of stemness-associated transcription factors. Sci Rep. 2018;8:7018.
Shaheen F, Hammad Aziz M, Fakhar EAM, Atif M, Fatima M, Ahmad R, Hanif A, Anwar S. An in vitro study of the photodynamic effectiveness of GO-ag nanocomposites against human breast Cancer cells. Nanomaterials (Basel). 2017;7:55–63.
Li P, Feng C, Chen H, Jiang Y, Cao F, Liu J, Liu P. Elevated CRB3 expression suppresses breast cancer stemness by inhibiting beta-catenin signalling to restore tamoxifen sensitivity. J Cell Mol Med. 2018;22:3423–33.
Lakota J. Fate of human mesenchymal stem cells (MSCs) in humans and rodents-is the current paradigm obtained on rodents applicable to humans? J Cell Mol Med. 2018;22:2523–4.
Yang S, Dong F, Li D, Sun H, Wu B, Sun T, Wang Y, Shen P, Ji F, Zhou D. Persistent distention of colon damages interstitial cells of Cajal through ca(2+) -ERK-AP-1-miR-34c-SCF deregulation. J Cell Mol Med. 2017;21:1881–92.
Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, Cui B, Zheng FM, Wang HJ, Lam EW, Wang B, Xu J, Liu Q. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569.
Su F, He W, Chen C, Liu M, Liu H, Xue F, Bi J, Xu D, Zhao Y, Huang J, Lin T, Jiang C. The long non-coding RNA FOXD2-AS1 promotes bladder cancer progression and recurrence through a positive feedback loop with Akt and E2F1. Cell Death Dis. 2018;9:233.
Fang Z, Gong C, Yu S, Zhou W, Hassan W, Li H, Wang X, Hu Y, Gu K, Chen X, Hong B, Bao Y, Chen X, Zhang X, Liu H. NFYB-induced high expression of E2F1 contributes to oxaliplatin resistance in colorectal cancer via the enhancement of CHK1 signaling. Cancer Lett. 2018;415:58–72.
Deng J, Yang M, Jiang R, An N, Wang X, Liu B. Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the Cancer stem-like cell (CSC) subpopulation enriched from breast Cancer cells. PLoS One. 2017;12:e0170860.
Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang X, Cai J. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685–92.
Acknowledgements
Not applicable.
Funding
This work was supported by National Science Foundation of China (81260071) and Guangxi Provincial Education Department Funding (KY2015YB232, KY2016YB342).
Availability of data and materials
The data in the current study are available from the corresponding authors on reasonable request.
Author information
Authors and Affiliations
Contributions
SH, ZW conceived and designed the study. GL, YL, YM performed the experiments and analyzed the data. JL, YC, QJ, QQ, LZ, QH, ZL assist the experiments. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki principles. It was approved by the Medical Research Ethics Committee of Youjiang Medical College Affiliated Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Sequences of shRNA and qRT-PCR. (DOCX 17 kb)
Additional file 2:
Table S2. Primers sequences for ChIP. (DOCX 16 kb)
Additional file 3:
Table S3. Nanog promoter region. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lu, G., Li, Y., Ma, Y. et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res 37, 289 (2018). https://doi.org/10.1186/s13046-018-0945-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13046-018-0945-6