Abstract
Background
Gastric cancer (GC) is one of the most common human cancers. Genes expressed only in cancer tissue, especially on the cell membrane, will be useful biomarkers for cancer diagnosis and therapeutics.
Methods
To identify novel genes encoding transmembrane protein specifically expressed in GC, we generated an Escherichia coli ampicillin secretion trap (CAST) library from diffuse-type GC cell line MKN-45. CAST is a survival-based signal sequence trap method that exploits the ability of mammalian signal sequences to confer ampicillin resistance to a mutant β-lactamase lacking the endogenous signal sequence.
Results
By sequencing 1,536 colonies, we identified 23 genes encoding the transmembrane protein present in GC. Among these genes, TSPAN8 (also known as CO-029 and TM4SF3) gene, which encodes transmembrane protein tetraspanin 8, was emphasized as a candidate. Immunohistochemical analysis of tetraspanin 8 in human GC tissues revealed that 72 (34 %) of 210 GC cases were positive for tetraspanin 8, and microvessel density was significantly higher in tetraspanin 8-positive GC than in tetraspanin 8-negative GC. Furthermore, univariate and multivariate analyses revealed that tetraspanin 8 expression is an independent prognostic classifier of patients with GC. TSPAN8 knockdown by siRNA reduced the invasion of GC cell line. The reduction of invasiveness was retrieved by the tetraspanin 8-containing exosome.
Conclusion
These results suggest that tetraspanin 8 is involved in tumor progression and is an independent prognostic classifier in patients with GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC), which develops as a result of multiple genetic and epigenetic alterations [1], is one of the most common human cancers. Better knowledge of the changes in gene expression that occur during gastric carcinogenesis may lead to improvements in the diagnosis, treatment, and prevention. The identification of biomarkers for cancer diagnosis and targets for treatment is a major goal in this field [2]. Genes that encode transmembrane/secretory proteins and that are expressed specifically in cancers can be ideal biomarkers for cancer diagnosis [3]. If the function of a gene product is involved in the neoplastic process, then the gene may constitute a therapeutic target.
It has been demonstrated that trastuzumab improves survival for patients with human epidermal growth factor receptor 2 (HER2)-positive advanced GC, which is frequently observed in intestinal-type GC [4]. In the case of diffuse-type GC, however, a novel therapeutic target is needed.
We previously performed an Escherichia coli ampicillin secretion trap (CAST) to identify transmembrane proteins present in GC cells [5]. CAST is a signal sequence trap method that was developed by Ferguson et al. [6]. Signal peptides, which typically consist of 4–15 hydrophobic amino acids flanked by a basic NH2 terminus and a polar COOH terminus, target secreted and transmembrane proteins in their appropriate subcellular location [7]. That a consensus sequence for the signal peptide has not been identified suggests that standard molecular techniques are not well suited to the identification of such proteins. CAST is a survival-based signal sequence trap method that exploits the ability of mammalian signal sequences to confer ampicillin resistance to a mutant β-lactamase, which lacks an endogenous signal sequence [8]. Using this method, we identified several GC-associated genes and reported that DSC2 was associated with the intestinal mucin phenotype of GC [5]. Although the authors confirmed the usefulness of CAST through a library from MKN-1 and MKN-28, they did not identify any GC-specific genes that were appropriate for use as novel biomarkers for cancer diagnosis or novel therapeutic targets.
In the present study we generated a CAST library from diffuse-type GC cell line MKN-45 to identify genes that encode transmembrane proteins present in GC, especially in diffuse-type GC. Secreted proteins were excluded from candidates in the present study because transmembrane proteins were considered to be better for therapeutics. Herein, we provide a list of genes that encode transmembrane proteins expressed in GC. Among these genes, we focused on the TSPAN8 (also known as CO-029 and TM4SF3) gene, which is overexpressed in GC and narrowly restricted in normal tissues. TSPAN8 encodes tetraspanin 8 protein, which is a member of the tetraspanins, a family of proteins that crosses the membrane four times and is involved in numerous biological processes [9]. In human cancers, overexpression of tetraspanin 8 has been reported in relation to hepatocellular carcinoma [10], pancreatic cancer [11], colon cancer [12], and esophageal cancer [13]. It has been also reported that TSPAN8 mRNA is upregulated in the GC tissues compared to normal gastric tissues by microarray analysis [14]. Furthermore, tetraspanins have been reported to be components of exosomes [15], and exosomes containing rat TSPAN8 have been shown to affect tumor cell migration, proliferation, and tumor angiogenesis [16]. In the present study, we examined the expression and distribution of tetraspanin 8 in human GC by immunohistochemistry and investigated the relationship between tetraspanin 8 staining and clinicopathologic characteristics. In addition, we investigated the biological function of tetraspanin 8 in cancer cell growth and invasiveness by RNA interference (RNAi). The role of tetraspanin 8-containing exosomes was also examined.
Materials and methods
Cell lines
Four GC cell lines were used (MKN-1, MKN-7, MKN-45, and MKN-74). All GC cell lines were purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). All cell lines were maintained in RPMI 1640 (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) containing 10 % fetal bovine serum (FBS, BioWhittaker, Walkersville, MD, USA) in a humidified atmosphere of 5 % CO2 and 95 % air at 37 °C.
CAST library construction
CAST library construction was performed as described previously [6].
Tissue samples
In total, 260 primary tumor samples were collected from patients diagnosed with GC who were treated at Hiroshima University Hospital or an affiliated hospital. This study was approved by the Ethics Committee for Human Genome Research of Hiroshima University.
For quantitative reverse transcription–polymerase chain reaction (qRT-PCR), 50 GC samples and corresponding non-neoplastic mucosa samples were used. Samples were frozen immediately in liquid nitrogen and stored at −80 °C until use. Fourteen types of normal tissue samples (heart, lung, stomach, small intestine, colon, liver, pancreas, kidney, bone marrow, leukocytes, spleen, skeletal muscle, brain, and spinal cord) were purchased from Clontech (Palo Alto, CA, USA).
For immunohistochemical analysis, archival formalin-fixed, paraffin-embedded tissues from 210 patients who had undergone surgical excision for GC were used. We examined one or two representative tumor blocks from each patient.
Tumor staging was determined according to the TNM classification system [17]. Histological classification of GC was determined according to the Lauren classification system [18].
Evaluation of the specificity of gene expression
To evaluate the specificity of the expression of each gene, a specificity index was calculated as described previously [19]. In brief, normal tissue in which the target gene expression was highest among the 14 types of normal tissues was identified then analyzed by qRT-PCR (the mRNA expression level in this tissue was denoted as A). Using qRT-PCR, we then identified GC among the nine GC samples in which the target gene expression was highest (the mRNA expression level in this tissue was denoted as B). The ratio of B to A was defined as the specificity index. When the specificity index of the target gene was ≥10, the gene was considered to show high specificity for GC. When the specificity index of the target gene was <10 and ≥2, the gene was considered to show low specificity for GC. When the specificity index of the target gene was <2, the gene was considered to show no specificity for GC.
Exosome preparation
TSAPN8 siRNA-trasfected MKN-45 cells and negative control siRNA-transfected MKN-45 cells were grown to 80–90 % confluence in RPMI 1640 medium containing 10 % FBS. The medium was removed, and the cells were washed twice with phosphate-buffered saline (PBS). Cells were incubated for 48 h in 20 ml serum-free RPMI 1640 medium. After 48 h, the medium was collected. After centrifugation of conditioned medium from TSAPN8 siRNA-transfected MKN-45 cells and negative control siRNA-transfected MKN-45 cells for 30 min at 3,000×g, supernatants were pelleted and washed in PBS at 100,000×g for 2 h and 10 min. The final pellet was resuspended in a small volume of PBS, and the protein content was measured using a DC protein assay in accordance with the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA, USA). Exosomes were aliquoted and frozen at −80 °C.
qRT-PCR, Western blot, immunohistochemistry, microvessel counts, RNAi, cell growth assay, in vitro invasion assay, wound-healing assay, and statistical methods were as follows.
qRT-PCR was performed with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) as described previously [20]. Western blot was performed as described previously [21]. RNAi was performed as described previously [22]. Cell growth was monitored by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [23]. These methods are described in detail in the Supplementary materials and methods.
Results
Generation of the MKN-45 CAST library
To identify transmembrane protein encoding genes, we generated a CAST library from diffuse-type GC cell line MKN-45 and sequenced 1,536 ampicillin–resistant colonies from the MKN-45 CAST library. The obtained sequences were compared with those deposited in public databases using BLAST (accessed at http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the subcellular localization of the gene products was evaluated using data from GeneCards (accessed at: http://www.genecards.org/index.shtml). From a total of 1,536 colonies, 1,210 were named human genes, which included 482 genes. In order for E. coli to survive the antibiotic challenge, the signal sequence and translation initiator ATG codon must be cloned in-frame with the leaderless β-lactamase reporter. Among the 482 genes, 237 were cloned in-frame upstream of the leaderless β-lactamase gene. Of these 237 genes, 10 encoded secreted proteins, 80 encoded transmembrane proteins, and 129 encoded proteins that were neither secreted nor transmembrane proteins. The remaining 18 genes have not been well studied.
Identification of genes expressed more highly in GC than in 14 types of normal tissue
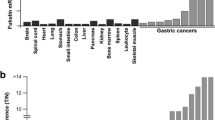
To identify genes expressed specifically in GC, we compared the gene list from the MKN-45 GC cell line CAST library to the normal stomach CAST library. The normal stomach CAST data had been previously determined [5]. Genes that were detected at least twice in the GC cell line CAST library, but not detected in the normal stomach CAST library, were selected. Finally, 23 candidates from MKN-45 were obtained (Table 1). To confirm that these candidates were GC-specific, qRT-PCR was performed to measure their expression in 9 GC tissue samples and in the 14 types of normal tissue. Representative data are shown in Fig. 1. Expression of the 23 candidates was not necessarily GC-specific; however, several showed much higher expression in GC than in normal tissues. We then focused on cancer specificity by calculating the specificity index (Table 1). Of the 23 candidates, 5 genes (ATP6AP2, SERINC3, TSPAN8, ATP11B, and SLC38A2) showed high specificity for GC (B/A ratio >10 and B/A ratio = 10); 11 genes (PLA2G2A, CDH17, SLC35F5, SLC12A2, TMEM41B, CLDN7, ITGB6, PLP2, MUC13, ZDHHC14, and TMCO3) showed low specificity for GC (B/A ratio <10 and B/A ratio >2).
qRT-PCR analysis of candidate genes that encode cell surface proteins in 14 types of normal tissues and 9 GC samples. Definitions of high specificity, low specificity, and no specificity are as described in “Materials and Methods.” mRNA expression levels of ATP6AP2, TSPAN8, and SLC38A2 were much higher in GC samples than in normal tissues. In contrast, mRNA expression levels of SLC12A6, CEACAM5, and TMEM50B did not differ significantly between GC and normal tissues
mRNA expression of TSPAN8 in GC and noncancerous tissues.
Among the five genes that showed high specificity for GC, we focused on TSPAN8 because it accounted for the largest number of detected clones in the MKN-45 CAST library. TSPAN8 encodes tetraspanin 8 protein, which is a member of the tetraspanin family. It has been reported that TSPAN8 is highly expressed in several types of human cancer [10–14]. Tetraspanins are involved in adhesion, motility, matrix modulation, and cell fusion [9]. Although it has been reported that TSPAN8 is expressed in the GC cell line [24], and that TSPAN8 mRNA is upregulated in GC tissues compared to normal gastric tissues according to microarray analysis [14], detailed expression of TSPAN8 has not been investigated. Among the 14 types of normal tissue, low-level TSPAN8 mRNA expression was detected in normal small intestine, colon, and liver. In normal gastric mucosa, TSPAN8 expression was extremely low (Fig. 1).
We analyzed TSPAN8 mRNA expression in 41 additional GC tissue samples and corresponding non-neoplastic mucosa samples. The ratios of TSPAN8 mRNA expression levels to GC tissue and corresponding non-neoplastic mucosa were calculated. A >2-fold ratio was considered to represent overexpression. TSPAN8 was overexpressed in 24 % (10/41) of the examined samples.
Immunohistochemical staining for tetraspanin 8 in GC and noncancerous tissues
To further analyze the expression of TSPAN8 in GC, we performed immunohistochemical analysis of tetraspanin 8. The mouse monoclonal anti-tetraspanin 8 antibody was detected in a single band of approximately 29 kDa on Western blot analysis of cell extracts from MKN-45 cells, suggesting that the anti-tetraspanin 8 antibody specifically recognizes tetraspanin 8 protein (data not shown). In qRT-PCR analysis, obvious mRNA expression was observed in normal small intestine and colon. Thus, immunohistochemical analysis was first performed in normal small intestine and colon to serve as positive controls. Staining of tetraspanin 8 was observed on cell membranes of the epithelial cells of the colon (Fig. 2a). Tetraspanin 8 staining was also detected on the cell membranes of the epithelial cells of the small intestine (data not shown).
Immunohistochemical analysis of tetraspanin 8 in non-neoplastic human tissues and GC tissues. a Immunostaining tetraspanin 8 in non-neoplastic colon revealed that colonic epithelial cells expressed tetraspanin 8 on cell membranes. Original magnification: ×200. b Immunostaining tetraspanin 8 in non-neoplastic gastric mucosa. Original magnification: ×200. c Immunostaining tetraspanin 8 in intestinal-type GC. Tetraspanin 8 staining was observed in GC cells on cell membranes. Original magnification: ×400. d Immunostaining tetraspanin 8 in diffuse-type GC. Tetraspanin 8 staining was observed in GC cells on cell membranes. Original magnification: ×400. e Immunostaining tetraspanin 8 in GC. In addition to GC cells, tetraspanin 8 staining showed capillary endothelial cells on cell membranes. Original magnification: ×400. Arrow, capillary blood vessel. Arrowhead, medium-sized blood vessel. Inset, capillary blood vessel. High magnification image of the fields indicated by the arrow is shown. f Kaplan-Meier plot of the survival in patients with GC
Next, immunohistochemical analysis was performed on 210 GC samples. Weak or no staining of tetraspanin 8 was observed in the epithelial cells in non-neoplastic gastric mucosa (Fig. 2b). Expression of tetraspanin 8 was not detected in inflammatory cells or fibroblasts. In contrast, GC tissues showed stronger, more extensive staining than corresponding non-neoplastic mucosa. Tetraspanin 8 staining was observed on cell membranes in both intestinal-type (Fig. 2c) and diffuse-type GC (Fig. 2d). In addition to cancer cells, capillary endothelial cells in GC tissue were also stained by tetraspanin 8 (Fig. 2e). Expression of tetraspanin 8 was found in both tetraspanin 8-positive and tetraspanin 8-negative GC cases. In contrast, tetraspanin 8 expression was not detected in endothelial cells of medium-sized blood vessels in GC tissue (Fig. 2e). In non-neoplastic gastric mucosa, tetraspanin 8 expression was not found in endothelial cells of capillaries or medium-sized blood vessels. In the present study, we only focused on tetraspanin 8 expression in cancer cells. When more than 10 % of cancer cell were stained, the immunostaining was considered positive for tetraspanin 8. In total, 72 (34 %) of 210 GC cases were positive for tetraspanin 8.
The relationship of tetraspanin 8 staining to clinicopathologic characteristics is shown in Table 2. No statistically significant relationship was found between tetraspanin 8 expression and any clinicopathologic features. However, the survival rate of patients who were GC positive for tetraspanin 8 was significantly worse than that of patients negative for tetraspanin 8 (P = 0.002, log-rank test, Fig. 2f). To evaluate the potential use of tetraspanin 8 expression as a prognostic classifier, univariate and multivariate Cox proportional hazards models were used to further evaluate the association between tetraspanin 8 expression and cancer-specific mortality (Table 3). In univariate analysis, the expression of tetraspanin 8 (HR 2.605; 95 % CI 1.371–4.952; P = 0.004) and tumor stage (HR 12.044; 95 % CI 5.756–27.746; P < 0.001) were associated with survival. In the multivariate model, which included tetraspanin 8 expression and tumor stage, tetraspanin 8 expression and tumor stage were found to be independent predictors of survival in patients with GC (Table 3).
It has been reported that tumor-derived exosomes containing tetraspanin 8 can induce angiogenesis in tumorous tissue [16]. Thus, we also analyzed the association between tetraspanin 8 expression and microvessel density. The microvessel density of tetraspanin 8-positive GC (mean ± SE, 47.5 ± 2.0) was significantly higher than that of tetraspanin 8-negative GC (mean ± SE, 41.0 ± 21.7, P = 0.019).
Effect of tetraspanin 8 inhibition on cell growth and invasive activity
Tetraspanin 8 has been reported to promote cell growth and migration through its cooperation with integrins and non-integrin partners [11, 25]. Since the biological impact of tetraspanin 8 in GC has not been investigated, we performed cell growth and in vitro invasion assays. MKN-45 cells, which showed the highest expression of tetraspanin 8 in the four GC cell lines, were selected for these experiments (Fig. 3a). To investigate the possible antiproliferative effects of TSPAN8 knockdown, we performed an MTT assay 8 days after siRNA transfection. Western blotting confirmed the suppressed expression of tetraspanin 8 by RNAi in MKN-45 cells (Fig. 3b). TSAPN8 siRNA1-trasfected and siRNA2-transfected MKN-45 cells showed significantly reduced cell growth relative to negative control siRNA-transfected MKN-45 cells (Fig. 3c).
Functional analysis of tetraspanin 8 in GC cell lines. a Western blot analysis of tetraspanin 8 in four GC cell lines. b Western blot analysis of tetraspanin 8 in cell lysates from MKN-45 cells transfected with TSPAN8 siRNA1, TSPAN8 siRNA2, and negative control siRNA. c Effect of TSPAN8 knockdown on cell growth of MKN-45 cells. Cell growth was assessed by an MTT assay at 1, 2, 4, and 8 days after seeding on 96-well plates. Bars and error bars, mean and SE of three different experiments. d Effect of TSPAN8 knockdown on cell invasion of MKN-45 cells. MKN-45 cells transfected with negative control siRNA, TSPAN8 siRNA1, and TSPAN8 siRNA2 were incubated in Boyden chambers. After 1 and 2 days, invading cells were counted. Bars and error bars, mean and SE of three different experiments. NS Not significant
To determine the possible role of tetraspanin 8 in the invasiveness of GC cells, we used modified Boyden chamber assays. On day 2, the invasiveness of TSAPN8 siRNA1-trasfected and siRNA2-transfected MKN-45 cells was less than that of the negative control siRNA-transfected MKN-45 cells (Fig. 3d). We also investigated cell migration activity by wound-healing assay. On fibronectin-coated culture slides, cell migration activity was inhibited in TSPAN8 siRNA1-trasnfected and siRNA2-transfected MKN-45 cells compared to negative control siRNA-transfected cells (data not shown). However, as TSPAN8 knockdown cells showed significantly reduced cell growth, the differences in cell number observed in the invasion assay and wound healing assay could be caused by the reduced cell growth. These results suggest that tetraspanin 8 stimulates cell growth and invasion in GC cancer cells.
Effect of tetraspanin 8 containing exosomes on cell growth and invasive activity
Exosomes, 50–100-nm vesicles, derived from endosomes forming multivesicular bodies, are released by various cells such as tumor cells, epithelial cells, dendritic cells, and lymphocytes. Exosomes contain selectively enriched mRNA and miRNA, which regulate gene expression in target cells. Tetraspanin 8 has been reported to be a component of exosomes [16]. We therefore examined the effect of tetraspanin 8-containing exosomes on GC cells. Western blot analysis revealed that exosome preparations of both TSAPN8 siRNA1-transfected MKN-45 cells (exosome-siTSPAN8-1) and negative control siRNA-transfected MKN-45 cells (exosome-NC) contained the exosome marker CD63 (Fig. 4a). We confirmed that tetraspanin 8 protein was reduced in exosome-siTSPAN8-1 compared to exosome-NC by Western blot.
Effect of tetraspanin 8-containing exosomes in GC cells. a Western blot analysis of CD63 and tetraspanin 8 in exosome preparations of both negative control siRNA-transfected MKN-45 cells (exosome-NC) and TSAPN8 siRNA1-transfected MKN-45 cells (exosome-siTSPAN8-1). b Effect of tetraspanin 8-containing exosomes in TSPAN8 siRNA1-transfected MKN-45 cells on cell growth. Cell growth was assessed by an MTT assay at 1, 2, 4, and 8 days after seeding on 96-well plates. Bars and error bars, mean and SE of three different experiments. c Effect of tetraspanin 8-containing exosomes in TSPAN8 siRNA1-transfected MKN-45 cells on cell invasion. MKN-45 cells transfected with TSPAN8 siRNA1 were incubated in Boyden chambers with exosome-NC or exosome-siTSPAN8-1. After 1 and 2 days, invading cells were counted. Bars and error bars, mean and SE of three different experiments. NS Not significant
We analyzed the effect of exosome preparations on cell growth in TSPAN8 siRNA1-transfected MKN-45 cells by MTT assay. The cell growth of TSPAN8 siRNA1-transfected MKN-45 cells treated with exosome-siTSPAN8-1 did not differ significantly from that of TSPAN8 siRNA1-transfected MKN-45 cells (Fig. 4b). In contrast, the cell growth of TSPAN8 siRNA1-transfected MKN-45 cells treated with exosome-NC was significantly higher than that of TSPAN8 siRNA1-transfected MKN-45 cells. We analyzed the effect of exosome preparations on cell growth in TSPAN8 siRNA2-transfected MKN-45 cells by MTT assay, and similar results were obtained (data not shown). These results suggest that tetraspanin 8-containing exosomes can promote GC cell growth.
We also analyzed the effect of exosome preparations on invasiveness in TSPAN8 siRNA1-transfected MKN-45 cells by a modified Boyden chamber assay. The invasiveness of TSPAN8 siRNA1-transfected MKN-45 cells treated with exosome-siTSPAN8-1 did not differ significantly from that of TSPAN8 siRNA1-transfected MKN-45 cells (Fig. 4c). In contrast, the invasiveness of TSPAN8 siRNA1-transfected MKN-45 cells treated with exosome-NC was significantly higher than that of TSPAN8 siRNA1-transfected MKN-45 cells. We analyzed the effect of exosome preparations on the invasiveness in TSPAN8 siRNA2-transfected MKN-45 cells by a modified Boyden chamber assay, and similar results were obtained (data not shown). These results suggest that tetraspanin 8-containing exosomes can promote GC cell invasion. However, because it is usual to use two cell lines as materials to show that the results are not specific to a cell line, but general, the effect of tetraspanin 8-containing exosomes on the growth and invasion of other GC cells should be examined in the near future.
Discussion
To identify genes that encode transmembrane proteins present in GC, especially in diffuse-type GC, we generated a CAST library from diffuse-type GC cell line MKN-45. We identified several genes that encode transmembrane proteins present in GC. qRT-PCR revealed that expressions of ATP6AP2, SERINC3, TSPAN8, ATP11B, and SLC38A2 were much higher in GC than in 14 types of normal tissues. Among these five genes, which showed high specificity for GC, we focused on TSPAN8, which encodes tetraspanin 8 protein. Tetraspanins are a family of proteins that cross the membrane four times and are involved in numerous biological processes [9]. Although the overexpression of tetraspanin 8 has been described in relation to several human cancers [10–13], and it has been also reported that TSPAN8 mRNA is upregulated in the GC tissues compared to the normal gastric tissues by microarray analysis [14], the significance of tetraspanin 8 protein in GC remains unclear. Immunohistochemical analysis revealed that tetraspanin 8 protein was expressed in GC cells and that 72 (34 %) of 210 GC cases were positive for tetraspanin 8. Furthermore, univariate and multivariate analyses demonstrated that tetraspanin 8 protein expression is an independent prognostic classifier of patients with GC. These results indicate that immunohistochemical analysis of tetraspanin 8 is a clinically useful method for the prediction of GC patient survival.
In the present study, tetraspanin 8 protein expression was shown to be an independent prognostic classifier for patients with GC. In addition, TSPAN8 knockdown cells were less invasive than negative control siRNA-transfected cells. TSPAN8 siRNA-transfected MKN-45 cells treated with exosome-NC were significantly more invasive than TSPAN8 siRNA-transfected MKN-45 cells treated with exosome-siTSPAN8. Furthermore, the microvessel density of tetraspanin 8-positive GC was significantly higher than that of tetraspanin 8-negative GC. These results indicate that tetraspanin 8 was involved in tumor progression rather than GC pathogenesis. Given that tetraspanin 8 is frequently overexpressed in GC and that it is narrowly restricted in normal tissues, it could be a good therapeutic target with fewer adverse effects for GC.
In addition to GC cells, tetraspanin 8 expression was observed in the capillary endothelial cells of GC tissue. It has previously been reported that capillary endothelial cells express tetraspanin 8 [26]. Interestingly, tetraspanin 8 expression was not observed in capillary endothelial cells in non-neoplastic gastric mucosa. The vascular anatomy of tumors is typically distinct from that of normal tissues and, using tetraspanin 8 conjugated to anti-tumor drugs, it may be possible to selectively target anti-tumor drugs to GC. E-cadherin, claudin, EPCAM, α6β4 integrin, CD44v, and EWI are suggested as key associated transmembrane proteins for TSPAN8, and antibodies against them may block tumor angiogenesis in treating patients with TSPAN8-positive tumors [27]. Actually, Ailane et al. [28] reported that in a nude mouse model, a monoclonal antibody against TSPAN8 inhibited the growth of tumors expressing TSPAN8 up to 70 % in vivo, while it did not affect the cell growth in vitro.
TSAPN8 siRNA-transfected MKN-45 cells showed significantly reduced cell growth in comparison to negative control siRNA-transfected MKN-45 cells. Although the underlying mechanisms remain unclear, it has been reported that tetraspanin 8 is associated with increased resistance to apoptosis [29]. The ability of tumor cell populations to expand in number is determined not only by the rate of cell proliferation, but also by the rate of cell attrition. Apoptosis represents a major source of this attrition. Because resistance to apoptosis contributes to chemotherapy resistance, the expression of tetraspanin 8 could be associated with chemotherapy efficiency. The association between tetraspanin 8 expression and chemotherapy resistance should be examined. Because TSPAN8 knockdown by siRNA may affect expression of other genes, overexpression of TSPAN8 should be performed in the near future.
In conclusion, our present study yielded a list of genes that encode transmembrane proteins present in GC. We found that tetraspanin 8 is expressed in GC cell membrane and that tetraspanin 8 expression is associated with poor survival. The purpose of this study is to identify genes that encode the transmembrane protein present in diffuse-type GC. Although tetraspanin 8 is equally expressed by both intestinal- and diffuse-type GC, tetraspanin 8-positive GC cases were observed in 34 % of diffuse-type GC. Because in diffuse-type GC only 3.4 % of cases are positive for HER2 [30], tetraspanin 8 could be a good therapeutic target for diffuse-type GC rather than HER2.
References
Yasui W, Sentani K, Sakamoto N, Anami K, Naito Y, Oue N. Molecular pathology of gastric cancer: research and practice. Pathol Res Pract. 2011;207:608–12.
Yasui W, Oue N, Ito R, Kuraoka K, Nakayama H. Search for new biomarkers of gastric cancer through serial analysis of gene expression and its clinical implications. Cancer Sci. 2004;95:385–92.
Buckhaults P, Rago C, St Croix B, Romans KE, Saha S, Zhang L, et al. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61:6996–7001.
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97.
Anami K, Oue N, Noguchi T, Sakamoto N, Sentani K, Hayashi T, et al. Search for transmembrane protein in gastric cancer by the Escherichia coli ampicillin secretion trap: expression of DSC2 in gastric cancer with intestinal phenotype. J Pathol. 2010;221:275–84.
Ferguson DA, Muenster MR, Zang Q, Spencer JA, Schageman JJ, Lian Y, et al. Selective identification of secreted and transmembrane breast cancer markers using Escherichia coli ampicillin secretion trap. Cancer Res. 2005;65:8209–17.
von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–90.
Kadonaga JT, Gautier AE, Straus DR, Charles AD, Edge MD, Knowles JR. The role of the beta-lactamase signal sequence in the secretion of proteins by Escherichia coli. J Biol Chem. 1984;259:2149–54.
Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55.
Kanetaka K, Sakamoto M, Yamamoto Y, Yamasaki S, Lanza F, Kanematsu T, et al. Overexpression of tetraspanin CO-029 in hepatocellular carcinoma. J Hepatol. 2001;35:637–42.
Gesierich S, Paret C, Hildebrand D, Weitz J, Zgraggen K, Schmitz-Winnenthal FH, et al. Colocalization of the tetraspanins, CO-029 and CD151, with integrins in human pancreatic adenocarcinoma: impact on cell motility. Clin Cancer Res. 2005;11:2840–52.
Greco C, Bralet MP, Ailane N, Dubart-Kupperschmitt A, Rubinstein E, Le Naour F, et al. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res. 2010;70:7674–83.
Zhou Z, Ran YL, Hu H, Pan J, Li ZF, Chen LZ, et al. TM4SF3 promotes esophageal carcinoma metastasis via upregulating ADAM12m expression. Clin Exp Metastasis. 2008;25:537–48.
Matsumura N, Zembutsu H, Yamaguchi K, Sasaki K, Tsuruma T, Nishidate T, et al. Identification of novel molecular markers for detection of gastric cancer cells in the peripheral blood circulation using genome-wide microarray analysis. Exp Ther Med. 2011;2:705–13.
Zöller M. Gastrointestinal tumors: metastasis and tetraspanins. Z Gastroenterol. 2006;44:573–86.
Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell surface tetraspanin TSPAN8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668–78.
Sobin LH, Compton CC. TNM seventh edition: what’s new, what’s changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–9.
Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49.
Aung PP, Oue N, Mitani Y, Nakayama H, Yoshida K, Noguchi T, et al. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene. 2006;25:2546–57.
Kondo T, Oue N, Yoshida K, Mitani Y, Naka K, Nakayama H, et al. Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer Res. 2004;64:523–9.
Yasui W, Ayhan A, Kitadai Y, Nishimura K, Yokozaki H, Ito H, et al. Increased expression of p34cdc2 and its kinase activity in human gastric and colonic carcinomas. Int J Cancer. 1993;53:36–41.
Oue N, Naito Y, Hayashi T, Takigahira M, Kawano-Nagatsuma A, Sentani K, et al. Signal peptidase complex 18, encoded by SEC11A, contributes to progression via TGF-alpha secretion in gastric cancer. Oncogene. 2014;33:3918–26.
Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601.
Sela BA, Steplewski Z, Koprowski H. Colon carcinoma-associated glycoproteins recognized by monoclonal antibodies CO-029 and GA22-2. Hybridoma. 1989;8:481–91.
Herlevsen M, Schmidt DS, Miyazaki K, Zöller M. The association of the tetraspanin D6.1A with the alpha6beta4 integrin supports cell motility and liver metastasis formation. J Cell Sci. 2003;116:4373–90.
Gesierich S, Berezovskiy I, Ryschich E, Zöller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083–94.
Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60.
Ailane N, Greco C, Zhu Y, Sala-Valdes M, Billard M, Casal I, et al. Effect of an anti-human Co-029/TSPAN8 mouse monoclonal antibody on tumor growth in a nude mouse model. Front Physiol. 2014;5:364.
Huerta S, Harris DM, Jazirehi A, Bonavida B, Elashoff D, Livingston EH, et al. Gene expression profile of metastatic colon cancer cells resistant to cisplatin-induced apoptosis. Int J Oncol. 2003;22:663–70.
Aizawa M, Nagatsuma AK, Kitada K, Kuwata T, Fujii S, Kinoshita T, et al. Evaluation of HER2-based biology in 1,006 cases of gastric cancer in a Japanese population. Gastric Cancer. 2014;17:34–42.
Acknowledgments
This work was supported in part by Grants-in-Aid for Cancer Research from the Ministry of Education, Culture, Sciences, Sports, and Technology of Japan and in part by a Grant-in-Aid for the Third Comprehensive 10-years Strategy for Cancer Control and for Cancer Research from the Ministry of Health, Labour, and Welfare of Japan.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anami, K., Oue, N., Noguchi, T. et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer 19, 370–380 (2016). https://doi.org/10.1007/s10120-015-0478-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-015-0478-z