Abstract
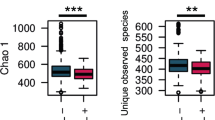
Methicillin-resistant Staphylococcus aureus (MRSA) may cause potentially lethal infections. Increasing evidence suggests that the gut microbiota is associated with human health. Yet, whether patients with MRSA infections carry specific signatures in their fecal microbiota composition has not been determined. Thus, this study aimed to compare the fecal microbiota profile of MRSA-positive patients (n=15) with individuals without MRSA infection (n=15) by using the PacBio single molecule, real-time (SMRT) DNA sequencing system and real-time quantitative polymerase chain reaction (qPCR). Mann-Whitney tests and unweighted UniFrac principal coordinate analysis (PCoA) showed that the profile of fecal microbiota was apparently different between the two populations. Both the community richness and diversity were reduced in the MRSA-positive group (p<0.050). The genera Acinetobacter and Enterococcus were highly enriched in the MRSA-positive group, whereas less short-chain fatty acid (SCFA)-producing bacteria, including Butyricimonas, Faecalibacterium, Roseburia, Ruminococcus, Megamonas and Phascolarctobacterium, were detected in the MRSA-positive group. At species level, the species Acinetobacter baumannii and Bacteroides thetaiotaomicron were prevalent in the MRSA-positive group, whereas opposite trends were observed in 17 other species, such as Faecalibacterium prausnitzii, Lactobacillus rogosae, Megamonas rupellensis and Phascolarctobacterium faecium. Positive correlations were observed between Acinetobacter baumannii and erythrocyte sedimentation rate (ESR) (R=0.554, p=0.001), as well as hypersensitive C reactive protein (hsCRP) (R=0.406, p=0.026). Faecalibacterium prausnitzii was negatively associated with ESR (R=-0.545, p=0.002), hsCRP (R=-0.401, p=0.028) and total bile acids (TBA) (R=-0.364, p=0.048). In conclusion, the fecal microbiota structure was different between MRSA-positive and -negative patients. The increase in potential pathogens with the reduction of beneficial populations, such as SCFA-producing bacteria, in MRSA-positive patients may affect prognosis.






Similar content being viewed by others
References
Qin N, Tan X, Jiao Y, Liu L, Zhao W, Yang S, Jia A (2013) RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep 4:5467–5467. doi:10.1038/srep05467
Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H (2012) Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. J Infect Dis 205(11):1688–1696. doi:10.1371/journal.pone.0041075
Thangamani S, Younis W, Seleem MN (2015) Repurposing ebselen for treatment of multidrug-resistant staphylococcal infections. Sci Rep 5. doi:10.1038/srep11596
Adamu BO, Lawley TD (2013) Bacteriotherapy for the treatment of intestinal dysbiosis caused by Clostridium difficile infection. Curr Opin Microbiol 16(5):596–601. doi:10.1016/j.mib.2013.06.009
Hartstra AV, Nieuwdorp M, Herrema H (2016) Interplay between gut microbiota, its metabolites and human metabolism: dissecting cause from consequence. Trends Food Sci Technol. doi:10.1016/j.tifs.2016.08.009
Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M (2013) Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5(193):3390–3396. doi:10.1126/scitranslmed.3006438
Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Wei J, Cai S, Zhao L (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6(2):320–329. doi:10.1038/ismej.2011.109
Wei Y, Gong J, Zhu W, Guo D, Gu L, Li N, Li J (2015) Fecal microbiota transplantation restores dysbiosis in patients with methicillin resistant Staphylococcus aureus enterocolitis. BMC Infect Dis 15(1):1–8. doi:10.1186/s12879-015-0973-1
Brahe LK, Chatelier EL, Prifti E, Pons N, Kennedy S, Hansen T, Pedersen O, Astrup A, Ehrlich SD, Larsen LH (2015) Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutrit Diabetes 5(6):1755–1758. doi:10.1038/nutd.2015.9
Carles U, Vanni B, Silvia C, Ana D, Toussaint NC, Michele E, Lauren L, Lilan L, Asia G, Daniel N (2013) Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 81(3):965–973. doi:10.1128/IAI.01197-12
Magrini L, Gagliano G, Travaglino F, Vetrone F, Marino R, Cardelli P, Salerno G, Di SS (2014) Comparison between white blood cell count, procalcitonin and C reactive protein as diagnostic and prognostic biomarkers of infection or sepsis in patients presenting to emergency department. Clin Chem Lab Med 52(10):1465–1472. doi:10.1515/cclm-2014-0210
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. doi:10.1038/nmeth.f.303
Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26(2):266–267(262). doi:10.1093/bioinformatics/btp636
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. doi:10.1093/bioinformatics/btq461
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen A, McGarrell DM, Bandela A, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35(suppl 1):D169–D172. doi:10.1093/nar/gkl889
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26(7):1641–1650. doi:10.1093/molbev/msp077
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al (2009) A core gut microbiome in obese and lean twins. Nature 457(7228):480–484. doi:10.1038/nature07540
Mckenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD (2008) The Macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. Plos Pathogens 4(2):20. doi:10.1371/journal.ppat.0040020
Mcmurtry VE, Gupta RW, Tran L, Blanchard EE, Penn D, Taylor CM, Ferris MJ (2014) Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome 3(1):1–8. doi:10.1186/s40168-015-0075-8
Sakamoto M, Takagaki A, Matsumoto K, Kato Y, Goto K, Benno Y (2009) Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ‘Porphyromonadaceae’ isolated from rat faeces. Int J Syst Evol Microbiol 59(Pt 7):1748–1753. doi:10.1099/ijs.0.007674-0
Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, Inatomi O, Bamba S, Andoh A, Sugimoto M (2016) Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion 93(1):59–65. doi:10.1159/000441768
Scupham AJ, Patton TG, Bent E, Bayles DO (2008) Comparison of the cecal microbiota of domestic and wild turkeys. Microb Ecol 56(2):322–331. doi:10.1007/s00248-007-9349-4
Yohei W, Fumiko N, Masami M (2012) Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol 78(2):511–518. doi:10.1128/AEM.06035-11
Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, Zhang X, Zhao L (2014) Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Scientific Reports 5. doi: 10.1038/srep14405
Sartor RB (2010) Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology 139(6):1816–1819. doi:10.1053/j.gastro.2010.10.036
Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D (2011) Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54(2):562–572. doi:10.1002/hep.24423
Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, Wang G, Wang F, Xu J, Cao H (2015) A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. Isme J Multidiscipl J Microb Ecol 9 (9). doi: 10.1038/ismej.2015.11
Wang F, Yu T, Huang G, Cai D, Liang X, Su H, Zhu Z, Li D, Yang Y, Shen P (2015) Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J Microbiol Biotechnol 25(8):1195–1204. doi:10.4014/jmb.1410.10014
Jangi S, Gandhi R, Cox LM, Li N, Glehn FV, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL (2016) Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 7:12015. doi:10.1038/ncomms12015
Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B (2014) Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 52(2):398–406. doi:10.1128/JCM.01500-13
Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB (2013) Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2(1629):e01202. doi:10.7554/eLife.01202
Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504(7480):2729–2732. doi:10.1038/nature13041
Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y (2008) Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29(4):637–649. doi:10.1016/j.immuni.2008.08.009
Steinway SN, Biggs MB, Jr LT, Papin JA, Albert R (2015) Inference of network dynamics and metabolic interactions in the gut microbiome. Plos Comput Biol 11 (6). doi:10.1371/journal.pcbi.1004338
Strati F, Cavalieri D, Albanese D, Felice CD, Donati C, Hayek J, Jousson O, Leoncini S, Pindo M, Renzi D (2016) Altered gut microbiota in Rett syndrome. Microbiome 4 (1). doi:10.1186/s40168-016-0185-y
Sekirov I, Russell SL, Antunes LC, Finlay BB (2010) Gut microbiota in health and disease. Physiol Rev 90(3):859–904. doi:10.1152/physrev.00045.2009
Vonschillde MA, Hörmannsperger G, Weiher M, Alpert CA, Hahne H, Bäuerl C, Vanhuynegem K, Steidler L, Hrncir T, Pérez-Martínez G (2012) Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11(4):387–396. doi:10.1016/j.chom.2012.02.006
Lăzureanu V, Poroșnicu M, Gândac C, Moisil T, Bădițoiu L, Laza R, Musta V, Crișan A, Marinescu AR (2016) Infection with Acinetobacter baumannii in an intensive care unit in the Western part of Romania. BMC Infect Dis 16(S1):23–28. doi:10.1186/s12879-016-1399-0
Georgescu M, Gheorghe I, Dudu A, Czobor I, Costache M, Cristea VC et al (2016) First report of OXA-72 producing Acinetobacter baumanniiin in Romania. New Microbes New Infect 13:87–88. doi:10.1016/j.nmni.2016.07.004
Haisma EM, Rietveld MH, De BA, van Dissel JT, El GA, Nibbering PH (2013) Inflammatory and antimicrobial responses to methicillin-resistant Staphylococcus aureus in an in vitro wound infection model. Plos One 8(12):1611–1614. doi:10.1371/journal.pone.0082800
Muñoz-Planillo R, Franchi L, Miller LS, Núñez G (2009) A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus-induced activation of the Nlrp3 inflammasome. J Immunol 183(6):3942–3948. doi:10.4049/jimmunol.0900729
Mcarthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De PG, Ejim L (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemotherapy 57(7):3348–3357. doi:10.1128/AAC.00419-13
Sommer MO, Dantas G, Church GM (2009) Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325(5944):1128–1131. doi:10.4161/viru.1.4.12010
Gosalbes MJ, Vallès Y, Jiménez-Hernández N, Balle C, Riva P, Miravet-Verde S, de Vries LE, Llop S, Agersø Y, Sørensen SJ (2015) High frequencies of antibiotic resistance genes in infants’ meconium and early fecal samples. J Dev Origins Health Dis 1(1):1–10. doi:10.1017/S2040174415001506
Ubukata K, Nonoguchi R, Matsuhashi M, Konno M (1989) Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol 171(5):2882–2885. doi:10.1128/jb.171.5.2882-2885.1989
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdezhumarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105(43):16731–16736. doi:10.1073/pnas.0804812105
Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdezhumarán LG, Pigneur B (2015) Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 194(5003):127–128. doi:10.1136/gutjnl-2014-307649
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
None
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study was approved by the Affiliated Hospital of Inner Mongolia Medical University.
Informed consent
All participants involved in this study gave their informed consent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Sample information (DOC 61.5 kb)
Table S2
Primer information for quantitative polymerase chain reaction and reference strains (DOC 31.5 kb)
Table S3
Alpha-diversity of samples (DOC 57 kb)
Fig. S1
(TIFF 1 mb)
Fig. S1
(a) Shannon-Wiener curves and (b) rarefaction curves of each sample. Red and blue lines represent the MRSA-positive and -negative samples, respectively
Rights and permissions
About this article
Cite this article
Zhao, J., Nian, L., Kwok, L.Y. et al. Reduction in fecal microbiota diversity and short-chain fatty acid producers in Methicillin-resistant Staphylococcus aureus infected individuals as revealed by PacBio single molecule, real-time sequencing technology. Eur J Clin Microbiol Infect Dis 36, 1463–1472 (2017). https://doi.org/10.1007/s10096-017-2955-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-017-2955-2