Abstract
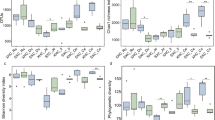
The extent to which production methods alter intestinal microbial communities of livestock is currently unknown. As the intestinal microbiota may affect animal health, nutrition, and food safety, a baseline comparison of the cecal communities of domestic and wild turkeys was performed. Oligonucleotide fingerprinting of ribosomal RNA (rRNA) genes (OFRG) of 2,990 16S rRNA clones and dot blot quantification of dominant populations were used to identify the dominant bacterial taxa. Seventy-three percent of all the clones belonged to as yet uncultured genera. However, at a higher phylogenetic level, the OFRG library was composed of 54% Bacteroidetes clones (52% of the domestic library clones, 56% of the wild library clones), 30% Firmicutes clones (33% of the domestic library clones, 32% of the wild library clones), 3% Proteobacteria clones (5% domestic, 2% wild), and 3% Deferribacteres clones (4% domestic, 1% wild). Seven percent of the clones were unidentifiable (6% domestic, 9% wild). Bacteroidetes clones included the genera Alistipes, Prevotella, Megamonas, and Bacteroides. Of the Clostridiales clones, groups IV, IX, and XIV including genera Faecalibacterium, Megasphaera, Phascolarctobacterium, and Papillibacter were predominant. Lactobacillus, Enterococcus, and Streptococcus bacilli were also identified. β- δ- and γ-proteobacterial genera included Acinetobacter, Sutterella, and Escherichia. Deferribacteres clones showed high similarity to Mucispirillum schaedleri. Statistical comparison of the domestic and wild turkey clone libraries indicated similar levels of community richness and evenness despite the fact that the two libraries shared only 30% of the total clone operational taxonomic units. Together these results indicate that although high level taxonomic community structure is similar, high-density turkey production causes considerable divergence of the genera found in the ceca of commercial birds from those of their wild counterparts.


Similar content being viewed by others
References
Amann R, Ludwig W (2000) Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol Rev 24:555–565
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amann RI, Krumholz L, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736
Barnes EM (1979) The intestinal microflora of poultry and game birds during life and after storage. Address of the president of the Society for Applied Bacteriology delivered at a meeting of the society on 10 January 1979. J Appl Bacteriol 46:407–419
Barnes EM, Impey CS, Stevens BJ (1979) Factors affecting the incidence and anti-salmonella activity of the anaerobic caecal flora of the young chick. J Hyg (Lond) 82:263–283
Benno Y, Endo K, Suzuki K, Mitsuoka T, Namioka S (1985) Use of nonprotein nitrogen in pigs: effects of dietary urea on the intestinal microflora. Am J Vet Res 46:959–962
Bent E, Yin B, Figueroa A, Ye X, Fu Q, Liu Z, Chrobak M, Jeske D, Jiang T, Borneman J (2006) Development of a 9,6000 clone array for oligonucleotide fingerprinting of rRNA genes: utilization to compare four different soil DNA extraction methods. J Microbiol Methods 67:171–180
Bielke LR, Elwood AL, Donoghue DJ, Donoghue AM, Newberry LA, Neighbor NK, Hargis BM (2003) Approach for selection of individual enteric bacteria for competitive exclusion in turkey poults. Poult Sci 82:1378–1382
Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J (1990) Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28:495–503
Borneman J, Chrobak M, Della Vedova G, Figueroa A, Jiang T (2001) Probe selection algorithms with applications in the analysis of microbial communities. Bioinformatics 17 Suppl 1:S39–S48
Coates JD, Cole KA, Michaelidou U, Patrick J, McInerney MJ, Achenbach LA (2005) Biological control of hog waste odor through stimulated microbial Fe(III) reduction. Appl Environ Microbiol 71:4728–4735
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:D294–D296
Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826
Dore J, Sghir A, Hannequart-Gramet G, Corthier G, Pochart P (1998) Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst Appl Microbiol 21:65–71
Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ (2002) Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52:2141–2146
Engberg J, On SL, Harrington CS, Gerner-Smidt P (2000) Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for Campylobacters. J Clin Microbiol 38:286–291
Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW (1998) Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 64:3336–3345
Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ (2006) Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol 44:4136–4141
Hanssen I (1979) A comparison of the microbiological conditions in the small intestine and caeca of wild and captive willow grouse (Lagopus lagopus lagopus). Acta Vet Scand 20:365–371
Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI (2001) Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884
Hughes JB, Bohannan BJM (2004) Application of ecological diversity statistics in microbial ecology. In: Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsasz JD (eds) Molecular microbial ecology manual, vol. 2, 2nd ed. Kluwer Academic Press, The Netherlands, pp 1321–1344
Hume ME, Kubena LF, Edrington TS, Donskey CJ, Moore RW, Ricke SC, Nisbet DJ (2003) Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult Sci 82:1100–1107
Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, Vogel U (2003) Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67–75
Jampachaisri K, Valinsky L, Borneman J, Press SJ (2005) Classification of oligonucleotide fingerprints: application for microbial community and gene expression analyses. Bioinformatics 21:3122–3130
Jernberg C, Lofmark S, Edlund C, Jansson JK (2007) Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The International Society for Microbial Ecology Journal 1:56–66
Jousimies-Somer H (1997) Recently described clinically important anaerobic bacteria: taxonomic aspects and update. Clin Infect Dis 25 Suppl 2:S78–S87
Krause DO, Smith WJ, Conlan LL, Gough JM, Williamson MA, McSweeney CS (2003) Diet influences the ecology of lactic acid bacteria and Escherichia coli along the digestive tract of cattle: neural networks and 16S rDNA. Microbiol 149:57–65
Lane DJ (1991) 16S/23S rRNA Sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824
Miroshnichenko ML, Bonch-Osmolovskaya EA (2006) Recent developments in the thermophilic microbiology of deep-sea hydrothermal vents. Extremophiles 10:85–96
Morotomi M, Nagai F, Sakon H (2007) Taxonomic note: genus Megamonas should be placed in the lineage of Firmicutes; Clostridia; Clostridiales; Acidaminococcaceae; Megamonas. Int J Syst Evol Microbiol 57:1673–1674
Nurmi E, Rantala M (1973) New aspects of Salmonella infection in broiler production. Nature 241:210–211
Pace NR, Stahl D, Lane DJ, Olsen GJ (1986) The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol 9:51–55
Raskin L, Stromley JM, Rittmann BE, Stahl DA (1994) Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60:1232–1240
Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A (2005) Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55:1199–1204
Salanitro JP, Blake IG, Muirhead PA (1974) Studies on the cecal microflora of commercial broiler chickens. Appl Microbiol 28:439–447
Schales K, Gerlach H, Kosters J (1993) Investigations on the aerobic flora and Clostridium perfringens in fecal specimens from free-living and captive capercaillies (Tetrao urogallus L., 1758). Zentralbl Veterinarmed B 40:469–477
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Handelsman J (2006) Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl Environ Microbiol 72:6773–6779
Scupham AJ (2007) Succession in the intestinal microbiota of preadolescent turkeys. FEMS Microbiol Ecol 60:136–147
Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J (2000) Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 66:2263–2266
Souza MR, Moreira JL, Barbosa FH, Cerqueira MM, Nunes AC, Nicoli JR (2007) Influence of intensive and extensive breeding on lactic acid bacteria isolated from Gallus gallus domesticus ceca. Vet Microbiol 120:142–150
Stern NJ, Svetoch EA, Eruslanov BV, Perelygin VV, Mitsevich EV, Mitsevich IP, Pokhilenko VD, Levchuk VP, Svetoch OE, Seal BS (2006) Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob Agents Chemother 50:3111–3116
Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Dore J (2001) Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 24:139–145
Valinsky L, Scupham AJ, Della Vedova J, Liu Z, Figuerosa A, Jampachaisri K, Yin B, Press J, Jiang T, Borneman J (2004) Oligonucleotide Fingerprinting of Ribosomal RNA Genes (OFRG). In Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD (eds) Molecular microbial ecology methods, vol. 1, 2nd ed. Kluwer Academic Press, The Netherlands, pp 569–585
Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ (2005) pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71:3692–3700
Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ (2006) Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40:235–243
Woodmansey EJ (2007) Intestinal bacteria and ageing. J Appl Microbiol 102:1178–1186
Yue JC, Clayton MK (2005) A similarity measure based on species proportions. Commun Stat 34:2123–2131
Yue JC, Clayton MK, Lin FC (2001) A nonparametric estimator of species overlap. Biometrics 57:743–749
Acknowledgments
The authors gratefully acknowledge the technical assistance of Jennifer A. Jones for all stages of data generation. We also thank David P. Alt and Karen Hollum for sequencing services.
Disclaimer
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
Dendrogram of the 2407 domestic and wild turkey clones and 39 control clones. The dendrogram was created using greedy clique partitioning of the oligonucleotide fingerprinting of rRNA genes (OFRG) fingerprints for each clone. Taxonomy for the labeled clones was determined by sequence analysis of the clone and RDP II classification of the sequence. Clone taxa appended with an asterisk (*) indicate taxonomy was determined by comparison of the fingerprint to theoretical fingerprints generated from GenBank sequence data (DOC 19 kb)
Rights and permissions
About this article
Cite this article
J Scupham, A., Patton, T.G., Bent, E. et al. Comparison of the Cecal Microbiota of Domestic and Wild Turkeys. Microb Ecol 56, 322–331 (2008). https://doi.org/10.1007/s00248-007-9349-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9349-4