Abstract
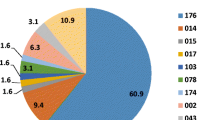
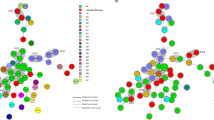
Clostridium difficile infection control strategies require an understanding of its epidemiology. In this study, we analysed the toxin genotypes of 130 non-duplicate clinical isolates of C. difficile from a university hospital in Tokyo, Japan. Multilocus sequence typing (MLST) and eBURST analysis were performed for these isolates and nine strains previously analysed by polymerase chain reaction (PCR) ribotyping. Minimum inhibitory concentrations (MICs) were determined for six antibiotics, and the bacterial resistance mechanisms were investigated. Ninety-five toxigenic strains (73 %), including seven tcdA-negative, tcdB-positive and cdtA/cdtB-negative strains (A−B+CDT−) and three A+B+CDT+ strains, and 35 (27 %) non-toxigenic strains, were classified into 23 and 12 sequence types, respectively. Of these, sequence type (ST)17 (21.8 %) was the most predominant. MLST and eBURST analysis showed that 139 strains belonged to seven groups and singletons, and most A+B+CDT− strains (98 %, 89/91) were classified into group 1. All isolates were susceptible to metronidazole, vancomycin and meropenem; the ceftriaxone, clindamycin and ciprofloxacin resistance rates were 49, 59 and 99 %, respectively. Resistance rates to ceftriaxone and clindamycin were higher in toxigenic strains than in non-toxigenic strains (P < 0.001). All ST17 and ST81 strains were resistant to these antibiotics. The clindamycin- and fluoroquinolone-resistant strains carried erm(B) and mutations in GyrA and/or GyrB, respectively. To our knowledge, this is the first MLST-based study of the molecular epidemiology of toxigenic and non-toxigenic strains in Japan, providing evidence that non-toxigenic and toxigenic strains exhibit high genetic diversity and that toxigenic strains are more likely than non-toxigenic strains to exhibit multidrug resistance.



Similar content being viewed by others
References
Depestel DD, Aronoff DM (2013) Epidemiology of Clostridium difficile infection. J Pharm Pract 26:464–475
George RH, Symonds JM, Dimock F, Brown JD, Arabi Y, Shinagawa N, Keighley MR, Alexander-Williams J, Burdon DW (1978) Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br Med J 1:695
Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y, Wasito EB (1998) Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol 36:2178–2182
Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084
Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ (2008) Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47:1162–1170
Sawabe E, Kato H, Osawa K, Chida T, Tojo N, Arakawa Y, Okamura N (2007) Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a five-year period. Eur J Clin Microbiol Infect Dis 26:695–703
Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJM, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE (2010) Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48:770–778
Kato H, Yokoyama T, Arakawa Y (2005) Typing by sequencing the slpA gene of Clostridium difficile strains causing multiple outbreaks in Japan. J Med Microbiol 54:167–171
Kato H, Kato N, Watanabe K, Yamamoto T, Suzuki K, Ishigo S, Kunihiro S, Nakamura I, Killgore GE, Nakamura S (2001) Analysis of Clostridium difficile isolates from nosocomial outbreaks at three hospitals in diverse areas of Japan. J Clin Microbiol 39:1391–1395
Oka K, Osaki T, Hanawa T, Kurata S, Okazaki M, Manzoku T, Takahashi M, Tanaka M, Taguchi H, Watanabe T, Inamatsu T, Kamiya S (2012) Molecular and microbiological characterization of Clostridium difficile isolates from single, relapse, and reinfection cases. J Clin Microbiol 50:915–921
Liao CH, Ko WC, Lu JJ, Hsueh PR (2012) Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother 56:3943–3949
Kim J, Kang JO, Pai H, Choi TY (2012) Association between PCR ribotypes and antimicrobial susceptibility among Clostridium difficile isolates from healthcare-associated infections in South Korea. Int J Antimicrob Agents 40:24–29
Weber I, Riera E, Déniz C, Pérez JL, Oliver A, Mena A (2013) Molecular epidemiology and resistance profiles of Clostridium difficile in a tertiary care hospital in Spain. Int J Med Microbiol 303:128–133
Persson S, Torpdahl M, Olsen KEP (2008) New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 14:1057–1064
Clinical and Laboratory Standards Institute (CLSI) (2012) Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. CLSI document M100-S22. CLSI, Wayne, PA
Nonaka S, Matsuzaki K, Kazama T, Nishiyama H, Ida Y, Koyano S, Sonobe K, Okamura N, Saito R (2014) Antimicrobial susceptibility and mechanisms of high-level macrolide resistance in clinical isolates of Moraxella nonliquefaciens. J Med Microbiol 63:242–247
Chung WO, Werckenthin C, Schwarz S, Roberts MC (1999) Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J Antimicrob Chemother 43:5–14
Drudy D, Quinn T, O’Mahony R, Kyne L, O’Gaora P, Fanning S (2006) High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother 58:1264–1267
Zilberberg MD, Shorr AF, Kollef MH (2008) Increase in adult Clostridium difficile-related hospitalizations and case–fatality rate, United States, 2000–2005. Emerg Infect Dis 14:929–931
Yan Q, Zhang J, Chen C, Zhou H, Du P, Cui Z, Cen R, Liu L, Li W, Cao B, Lu J, Cheng Y (2013) Multilocus sequence typing (MLST) analysis of 104 Clostridium difficile strains isolated from China. Epidemiol Infect 141:195–199
Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ (2009) Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of Toxin B or Toxin A. BMC Infect Dis 9:103
Collins DA, Hawkey PM, Riley TV (2013) Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2:21
Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ; ECDIS Study Group (2011) Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73
Spigaglia P, Barbanti F, Dionisi AM, Mastrantonio P (2010) Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J Clin Microbiol 48:2892–2896
Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, Kuijper EJ, Corver J, van Leeuwen HC (2012) Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol 12:1577–1585
Barbut F, Mastrantonio P, Delmée M, Brazier J, Kuijper E, Poxton I; European Study Group on Clostridium difficile (ESGCD) (2007) Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect 13:1048–1057
Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Courvalin P (1990) Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res 18:3660
Gregory ST, Dahlberg AE (1999) Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol 289:827–834
Hooper DC, Wolfson JS, Ng EY, Swartz MN (1987) Mechanisms of action of and resistance to ciprofloxacin. Am J Med 82:12–20
Spigaglia P, Barbanti F, Mastrantonio P, Brazier JS, Barbut F, Delmée M, Kuijper E, Poxton IR; European Study Group on Clostridium difficile (ESGCD) (2008) Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol 57:784–789
Walkty A, Boyd DA, Gravel D, Hutchinson J, McGeer A, Moore D, Simor A, Suh K, Taylor G, Miller M, Mulvey MR; Canadian Nosocomial Infection Surveillance Program (2010) Molecular characterization of moxifloxacin resistance from Canadian Clostridium difficile clinical isolates. Diagn Microbiol Infect Dis 66:419–424
Conflict of interest
The study did not receive financial support from any third party. All authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kuwata, Y., Tanimoto, S., Sawabe, E. et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis 34, 763–772 (2015). https://doi.org/10.1007/s10096-014-2290-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2290-9