Abstract
The quantitative mapping of food web flows based on empirical data is a crucial yet difficult task in ecology. The difficulty arises from the under-sampling of food webs, because most data sets are incomplete and uncertain. In this article, we review methods to quantify food web flows based on empirical data using linear inverse models (LIM). The food web in a LIM is described as a linear function of its flows, which are estimated from empirical data by inverse modeling. The under-sampling of food webs implies that infinitely many different solutions exist that are consistent with a given data set. The existing approaches to food web LIM select a single solution from this infinite set by invoking additional assumptions: either a specific selection criterion that has no solid ecological basis is used or the data set is artificially upgraded by assigning fixed values to, for example, physiological parameters. Here, we advance a likelihood approach (LA) that follows a different solution philosophy. Rather than singling out one particular solution, the LA generates a large set of possible solutions from which the marginal probability density function (mPDF) of each flow and correlations between flows can be derived. The LA is exemplified with an example model of a soil food web and is made available in the open-source R-software. Moreover, we show how stoichiometric data, stable isotope signatures, and fatty acid compositions can be included in the LIM to alleviate the under-sampling problem. Overall, LIM prove to be a powerful tool in food web research, which can bridge the gap between empirical data and the analysis of food web structures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The food web concept forms a cornerstone of modern ecology as it describes the exchange of matter—the so-called food web flows—among different compartments within an ecosystem. Early food web theory focused primarily on the topology of food webs, in which interactions among compartments are simply marked as either present or absent (Pimm and others 1991). In recent decades, ecologists have realized that a topological perspective on food web flows is insufficient to understand natural food webs and that one also needs to account for the magnitude of the flows, which is one measure of “interaction strength” in the ecological literature (Berlow and others 2004). Analysis of interaction strengths in natural food webs has revealed important features of their functioning. For example, the stability of soil food webs critically depends on the patterning of interaction strengths in the food web (De Ruiter and others 1995). A dominance of weak interactions in long trophic loops dampens the potentially destabilizing effect of such long loops (Neutel and others 2002). Moreover, the coupling of fast and slow trophic pathways by top predators also increases food web stability (Rooney and others 2006).
Ecological theory aims to explain various food web properties, such as the level of omnivory, food chain length, connectance, and the number of basal, intermediate and top species (for example, Williams and Martinez 2000). These food web descriptors are typically calculated from topological food webs. However, Banašek-Richter and others (2004) show that descriptors based on quantified food webs are superior to those based on topological food webs: fewer observations were needed to reliably constrain the descriptors and therefore better represented the food web structure. Accordingly, a vital step in food web research is to develop a systematic and standardized method to produce quantified food webs, so one can fully explore their structure and properties (Woodward and others 2005).
The problem of food web reconstruction basically comes down to finding the “best” (or most likely) set of flow values, assuming a certain food web topology and given an empirical data set. Historically, this problem was solved using a procedure of sequential mass-balancing: one postulates a certain food web topology and uses the available empirical data to close the mass balance of the top predator. The flow values from the predator’s mass balance are then used in the mass balances of their prey, and so on. This way, one can “mass balance down the food web” until the basal resources are resolved (for example, Hunt and others 1987). A clear advantage of this sequential mass-balancing is its simplicity, but there are also downsides. One problem is that the approach is single-currency: mass balances are expressed in a single element, typically either carbon (C) or nitrogen (N), and thus stoichiometric coupling is ignored. A more fundamental problem is the subjective nature of the approach. Field measurements on flows in food webs are difficult and laborious to obtain, and as a result, data sets are usually “incomplete.” The direct consequence is that the problem of food web reconstruction has no single “best” solution, but an infinite number of flow patterns that comply with the given data set. In mathematical terms, the problem is said to be under-determined. Because there is more than one possibility to close the mass balance of a food web compartment, subjective choices by the ecologist are needed to close the mass balances. These choices will determine the food web structure that is finally selected. When fewer data are available for lower trophic levels, it is there where errors eventually accumulate. Despite these disadvantages, the sequential mass-balancing procedure is still used in modern food web research (for example, Woodward and others 2005).
The ECOPATH software offers a more systematic approach to the mass-balancing problem (Christensen and Pauly 1992). A clear benefit of ECOPATH is that data input and mass-balancing are performed in a standardized and user-friendly way. More importantly, the problem of food web quantification is formulated in a rigorous mathematical way as a linear inverse model (LIM). This implies that the available food web data are parsed into a matrix equation, and the unknown flows are obtained by solving this linear equation system. Accordingly, mass balances are all solved simultaneously rather than sequentially. Nonetheless, ECOPATH remains restricted to single-currency data, as it does not allow the simultaneous solution of mass balances for multiple elements. Moreover, ECOPATH circumvents the problem of mathematical indeterminacy in a rather ad-hoc fashion. The number of equations is artificially upgraded until the matrix equation is completely determined by imposing fixed values for, for example, physiological parameters (see “Conversion efficiencies”). This again introduces a subjective aspect, because the researcher has to make choices on which additional data to include.
Food web studies have adopted two approaches to tackle the problem of data scarcity and model indeterminacy. Firstly, an improved LIM procedure has been proposed in the field of marine ecology. This LIM procedure solves multiple mass balances simultaneously and directly deals with the underdetermined matrix equation (Klepper and Van de Kamer 1987; Vézina and Platt 1988). Rather than artificially upgrading the data set, one uses the incomplete and/or uncertain data set as such, and selects a “best” solution from the infinite set of food web structures. The selection of this “best” solution is based on an optimization criterion (discussed in detail below). This improved LIM procedure has been used quite frequently in aquatic plankton ecology, but has found limited application in the wider ecological literature. One obstacle is presumably the absence of appropriate software to perform the required numerical calculations. Another issue is the question of whether the optimization criterion selects the “best” food web structure (Kones and others 2006). In other words, ecological theory does not provide a generally accepted “goal function” for food web functioning, which reliably selects a “best” food web structure from the infinite set of solutions.
A second way to deal with the problem of incomplete data and mathematical indeterminacy is to simply enlarge the empirical data set. Effectively, two novel data types have found widespread application in ecology that can significantly reduce the under-sampling of food webs. Firstly, important insights have been gained from ecological stoichiometry, in which food web interactions are constrained by the stoichiometric (im)balance among abiotic and biotic compartments (Sterner and Elser 2002). For example, Gaedke and others (2002) showed that zooplankton in Lake Constance acquired C through herbivory and phosphorous (P) by bacterivory, an interplay that fostered omnivorous pathways in the food web. Secondly, stable isotope data (13C and 15N) have provided important constraints on diet composition and trophic position of organisms under field conditions (see Minagawa and Wada 1984; Post 2002). These new categories of data provide additional mass balance constraints, provided that one can solve multiple mass balances (for example, C, N, P, 13C, and 15N) simultaneously. Only a LIM allows for such simultaneous solution of multiple mass balances.
In this article, we discuss two ways to improve the quantitative reconstruction of food webs using LIM. Firstly, we present a novel solution procedure for the underdetermined matrix equation system. Instead of selecting a single solution from the infinite set of solutions, the approach here uses a sampling method to retrieve the distribution of flow values in the solution set. A “best” flow value and its associated uncertainty can be inferred from all sampled food web solutions. This solution procedure is illustrated with an example soil food web model. The software used is made publically available through the packages LIM (Soetaert and Van Oevelen 2008) and limSolve (Soetaert and others 2008) that run in the R-software (R Development Core Team 2008). Secondly, we show how novel data types, such as stoichiometric and stable isotope data, can be implemented in a LIM. In this way, one can reduce the under-sampling problem and improve the quality of the food web reconstruction.
Linear Inverse Modeling: Model Formulation and Ecological Data Types
A LIM is termed linear because the food web model is described as a linear function of the flows. Inverse modeling means that model parameters are derived from observed data. Fitting a straight line through data points is a classical example of inverse modeling. In the context of this article, the magnitudes of the flows are the unknown model parameters that need to be quantified by fitting against an incomplete data set. The model itself is the topology of the food web, which is determined a priori by fixing the number of compartments and connecting these compartments with flows. The LIM incorporates the mass balance(s) of each compartment and a set of quantitative data constraints (as discussed in detail below). The overall structure of a LIM is formed by two matrix equations:
The vector x contains the unknown flows (that is, x 1, …, x n ), whereas the vectors f and h contain various types of empirical data. Each row in the equality equation (1) imposes a “hard” constraint: a linear combination of the flows must match the corresponding value in vector f. Therefore, equality constraints are used to incorporate high-quality data in the LIM, such as empirical data that are obtained from in situ sampling of the food web under study. Less strict data constraints are included via the inequality equation (2), where each row imposes a lower bound value on a linear combination of flows. This option is used for “soft” data constraints that are typically based on data that originate from elsewhere, for example, literature sources on other but comparable food webs. The inequality equation (2) appears to accept only lower bounds, but upper bound constraints can be implemented after converting them to lower bound constraints through multiplication of the left- and right-hand side with −1. A default set of inequalities is that x ≥ 0, which insures that flows have directions that are consistent with the food web topology (for example, predators can eat prey, but not the other way around).
There exists an overwhelming variety of empirical ecological data, which can be harnessed into the data vectors f and h. We distinguish these data based on “source” and “type.” The source of data relates to their origin; data are either directly obtained from the food web under study or from literature sources on other food webs. In general, we rank site-specific data as “high quality,” because they are obtained from the food web under study. Literature data are considered to be of lower quality, because they are usually derived from other food webs, and hence, it is not certain whether such data apply to the food web at hand.
The data type refers to the kind of data that is available and we will now briefly discuss six common data types.
Mass balances An essential physical constraint is that mass conservation should hold for each element. Each mass balance has the form: \( {{dx} \mathord{\left/ {\vphantom {{dx} {dt}}} \right. \kern-\nulldelimiterspace} {dt}} = \sum {x_{\text{in}} - \sum {x_{\text{out}} } } , \) stating that the temporal change in mass of compartment x should equal the difference between the incoming and outgoing flows. Data on temporal stock changes (dx/dt, for example, obtained by sampling stock sizes at consecutive times) are entered in the appropriate position in vector f (see the “MB” coded equations of the example model in Table 1). In case such data are not available, and one can assume steady-state and set the corresponding f i to zero.
Food web flows Specific measurements on the magnitude of an individual flow (for example, primary production) or a combination of several flows (for example, community respiration) provide direct constraints that can be directly implemented in the food web LIM equations (see “F” equation in Table 1). Such measurements are often difficult to conduct in situ, and usually only a limited amount of such data is available.
Biomass data are relatively easy to collect and form a cornerstone of quantitative food web research. Biomass data cannot be implemented directly, because the LIM is written in terms of flows rather than biomass. However, in combination with biomass-specific rate constants, biomass data provide important constraints on the flow magnitudes (see “BIO” equations in Table 1). Typical examples are biomass-specific growth, consumption, respiration, and mortality rates. Such rate constants are typically derived from laboratory experiments, and hence, their applicability to field conditions is uncertain. Accordingly, such data should generally be regarded as low quality.
Conversion efficiencies provide constraints dictated by the physiology of the organism or functional group in the food web. Classical examples are the assimilation efficiency, that is, the ratio of assimilated food over ingested food, and the growth efficiency, that is, the ratio of secondary production over assimilated food. Conversion efficiencies constrain the magnitude of flows relative to others (see “CE” equations in Table 1). Some conversion efficiencies can be estimated in situ, for example, assimilation efficiency from the Conover-ratio, that is, the change of the organic fraction of food during digestion (Conover 1966), and can then be considered as high-quality data. Other efficiencies, for example, growth efficiencies, are classically derived from laboratory experiments and should therefore be considered as low quality data.
Stoichiometry The stoichiometric composition of organisms in terms of C, N, and P is relatively easy to determine for organisms collected in the field. Like conversion efficiencies, such stoichiometry data (typically C:N, C:P, and N:P ratios) couple different element flows (see “STOI” equations in Table 1). In addition, stoichiometry constrains processes at the physiological, population, and ecosystem level (Sterner and Elser 2002). Such correlations can be used to constrain amongst others conversion efficiencies, growth rates, and/or decomposition rates directly (see “Discussion”).
Stable isotope signatures are used in ecology to decipher the relative importance of resources. The isotope signature of a consumer \( \left( {{{\updelta}}X_{j} } \right) \) is modeled as a weighed function of the isotope signature of its food sources \( \left( {{{\updelta}}X_{i} } \right), \) fractionation during trophic transfer \( \left( {\Updelta_{ij} } \right), \) and the relative contributions (α i ) of the food sources in the diet. This leads to a simple linear mixing model (for example, Phillips and Gregg 2003): \( {{\updelta}}X_{j} = \sum\limits_{i} {\alpha_{i} \left( {\delta X_{i} + \Updelta_{j} } \right),} \quad {\text{with }}\sum {\alpha_{i} } = 1. \) Because \( \alpha_{i} = {{{\text{flow}}_{i \to j} } \mathord{\left/ {\vphantom {{{\text{flow}}_{i \to j} } {\left( {\sum\limits_{i} {{\text{flow}}_{i \to j} } } \right)}}} \right. \kern-\nulldelimiterspace} {\left( {\sum\limits_{i} {{\text{flow}}_{i \to j} } } \right)}} \) this model can be directly rewritten as a linear function of the flows \( \left( {{\text{flow}}_{i \to j} } \right)\hbox{: }\delta X_{j} = {\frac{{\sum\nolimits_{i} {{\text{flow}}_{i \to j} \left( {\delta X_{i} + \Updelta_{j} } \right)} }}{{\sum\nolimits_{i} {{\text{flow}}_{i \to j} } }}}. \) The latter equation can be implemented in the equality equation (1) in a straightforward way (for example, Van Oevelen and others 2006).
Linear Inverse Modeling: Three Solution Approaches
When all data types are incorporated, the LIM equation system becomes
where n denotes the number of flows, m the number of mass balances, d the number of equalities, and c the number of inequalities. Equations (3) and (4) form the heart of the LIM, and a suitable solution procedure is required to recover the flow values in x. A key mathematical property of a LIM is the rank parameter r, which denotes the number of linearly independent equations in the matrix equation (3). When all equalities are linearly independent, the rank parameter r simply equals m + d.
When the number of independent equations matches the number of unknown flows (r = n = m + d), the matrix E is square and said to be of full rank. When there are no inequality constraints, the full-rank LIM has a unique solution, which can be directly found by matrix inversion: x = E −1 · f. This is analogous to fitting a straight line through two points. However, a full-rank LIM is never met in practice, because food webs are typically under-sampled (r < n). Moreover, inequality constraints are always present, because it is necessary to impose that x ≥ 0 to give flows a specific direction.
A LIM is said to be over-determined when the number of independent equations is larger than the number of flows (r > n). The analogous situation is fitting a straight line through more than two data points. In this case, the equality matrix equation (3) has no obvious solution, because not all equality constraints can be satisfied simultaneously. Instead of reproducing the equality constraints exactly, one seeks the solution that reproduces the equalities as well as possible (see below).
As noted above, food webs are typically under-sampled and the number of equalities is then insufficient to balance the number of unknown flows (r < n). In this case, the problem is said to be under-determined and the equality matrix equation (3) has an infinite number of solutions. The analogy is trying to fit a line through a single point; an infinite number of lines can be drawn. The main challenge in food web reconstruction is to solve this under-determinacy problem. In the next paragraphs, we discuss three ways to achieve this. The first two are well known from the literature, whereas the last one is a novel contribution.
Single-Solution Approach to an Over-Determined LIM
As discussed above, one usually makes a distinction between two kinds of data: (1) high-quality data that are incorporated as “hard” equality constraints (for example, site-specific data), and (2) lower quality data that are incorporated as “soft” inequality constraints (for example, literature data). One way to deal with under-determinacy is to make no distinction between data qualities and to treat site-specific and literature data equally. In other words, lower quality data from the literature are upgraded from soft inequalities to be incorporated as fixed “hard” equalities. After upgrading sufficient literature data to hard equalities, the number of equalities will equal and then exceed the number of flows (m + d > n), at which point the LIM has become over-determined. The single “best” solution to such an over-determined LIM solved is considered the flow vector x that minimizes the difference between the model prediction (E · x) and the data (f), raised to some power L:
The power L influences the weighting in the minimization and can have values of 1, 2, or ∞ (for example, Menke 1984): when L = 1, all differences are weighted equally, whereas larger values of L give comparatively more weight to large differences. This approach will henceforth be referred to as the single-solution approach to an over-determined LIM (SSAover) (see for example, Klepper and Van de Kamer 1987; Diffendorfer and others 2001; Gaedke and others 2002 for examples).
Single-Solution Approach to an Under-Determined LIM
In an alternative approach, one explicitly differentiates between equalities and inequalities as dictated by the data quality. In this case, one includes only site-specific data as “hard” equalities and literature data as “soft” inequalities. As stated above, the resulting under-determined LIM has infinitely many solutions that are all equally likely from a data perspective. It is therefore necessary to make an additional assumption to single out one “best” solution. One popular method is to select the “best” food web structure on the assumption of parsimony or simplicity. The most parsimonious solution is defined as the flow vector x that has the smallest sum of squared flow values (sensu Vézina and Platt 1988):
where L can again have values of 1, 2, or \( \infty. \) In most food web applications, L = 2 (sensu Vézina and Platt 1988), which requires that the sum of squared flows should be as small as possible and that flows should be partitioned as uniformly as possible (Niquil and others 1998). We will henceforth refer to this approach as the single-solution approach to the under-determined LIM (SSAunder) (see Vézina and Platt 1988; Jackson and Eldridge 1992; Richardson and others 2003 for examples). Although only: one single solution is selected in SSAunder, it is interesting to find the minimum and maximum values of each flow that exists in solution space, because it informs one on the upper and lower bounds in-between where the “best” flow value lies. These extremes can be found via a range estimation procedure: each flow is successively minimized and maximized under the condition that E · x = f and G · x ≥ h (Klepper and Van de Kamer 1987).
We see two potential downsides of SSAunder. Firstly, there is no theoretical or empirical evidence that robustly underpins the assumption of parsimony (Steele 2009). Therefore, we cannot be sure that the selected food web structure is correct. Secondly, the SSAunder can have unlikely properties: (1) some flows may be set to zero (Vézina and others 2004; Kones and others 2006) and (2) many flows are close to the lower bound of their ranges, which should be considered extreme values rather than likely ones (Diffendorfer and others 2001; Kones and others 2006; Steele 2009). Some of these disadvantages can be partially alleviated by adding additional smoothing among the flow values (Vézina and others 2004).
Likelihood Approach
We have shown above that both the SSAover and SSAunder require additional assumptions to deal with the under-determinacy of the LIM. Here, we discuss an alternative likelihood approach (LA) that does not need such additional assumptions. In the LA, we focus on “all” potential LIM solutions (that is, food web structures), rather than invoking additional assumptions to select a single food web. The distribution of all LIM solutions is formally captured in a probability density function (PDF). The distribution of values for each individual flow in all solutions is the marginal probability density function (mPDF), that is, the smoothed version of a histogram, and is obtained through integration of the PDF over the solution domain. The LA has four important advantages: (1) it is “objective” in the sense that no selection criterion is used, (2) low-quality data do not need to be implemented as high-quality data, (3) it solves both under- and over-determined LIM with the same methodology, and (4) information about the “quality” of the model inference can be obtained.
The LA is basically the same for under- and over-determined LIM, but it is easier to first detail the LA for under-determined models (LAunder). Figure 1A shows an under-determined LIM that for illustrative purposes involves only two flows. All combinations of values for flow1 and flow2 that are valid solutions to the LIM are enclosed in the grey region, whereas the invalid solutions are in the white area. The grey shading is uniform to emphasize that all solutions are equally likely from a “data-perspective”: the PDF of all solutions is constant over this area. The mPDF for flow1 is simply the integration of the surface area of the grey region over flow2 (Figure 1B). This gives an interesting result: although the PDF of all solutions is uniform, the mPDF of a flow is not uniform. Rather than trying to calculate the mPDF analytically, which is an arduous task for more complex LIM, it is possible to approximate the mPDF numerically by a Markov Chain Monte Carlo algorithm. In particular, we use the sampling algorithm xsample (Van den Meersche and others 2009) that is included in the R-package limSolve (Soetaert and others 2008). This sampling algorithm generates a set of consistent solutions (approximately thousands of solutions), from which the mPDF for each of the flows can be derived.
Schematic representation of the LA for an under-determined LIM. The grey region enclosed in equality constraints contains all combinations of flow values for flow1 and flow2 that are consistent with the data. The grey shading is uniform to signify that the PDF of all enclosed solutions is uniform. Solutions in the white region are inconsistent with the data constraints, and hence, their likelihood is zero. The sampling algorithm locates solutions (numbered dots) by taking random jumps (for example, from solutions 1 to 2). When the algorithm jumps out of the grey region, a reflection method mirrors the jump back into the grey region. The mPDF’s for the flow1 and flow2 are derived from the total set of sampled solutions. See “Linear inverse modeling: three solution approaches”–“Likelihood approach” for more details.
After the construction of the mPDF, the next step is to extract a single solution from the ensemble, for which different options are possible. The median flow values in the solution set give central estimates and the 25 and 75% quantiles indicate uncertainty of this central estimate. Alternatively, the maximum likelihood of each flow (that is, the value for which the mPDF is maximal) can be inferred using kernel density estimation on the set of sampled solutions (for example, by the R-function “density”). Both these approaches give a single “best” value for each flow, but neither the medians nor maximum likelihoods necessarily represent valid solutions to the LIM equations (3) and (4). The median and maximum likelihoods are valid solutions in the schematic example of Figure 1, but in more complex LIM this is not guaranteed. However, one can show that the mean flow vector always represents a valid solution to the LIM. This is because the calculation of the mean involves a linear operation on the solution set, which in turn gives a valid solution. This can be simply shown as follows. Consider a set of N vectors x N , that are all valid solutions to the LIM such that \( {\mathbf{E}} \cdot {\mathbf{x}}_{i} = {\mathbf{f}} \) with i = 1, …, N. The mean flow vector of the solution set is again a valid solution because \( {\mathbf{E}} \cdot \left( {{\frac{1}{N}} \cdot \sum {{\mathbf{x}}_{N} } } \right) = {\frac{1}{N}} \cdot \sum {\left( {{\mathbf{E}} \cdot {\mathbf{x}}_{N} } \right)} = {\frac{1}{N}} \cdot \left( {N \cdot {\mathbf{f}}} \right) = {\mathbf{f}} \) (a similar reasoning holds true for \( {\mathbf{Gx}} \ge {\mathbf{h}}). \) The example model discussed below shows that the mean flow values always closely approximate the medians, and therefore, the mean flow vector seems to provide a good central estimate that is consistent with the LIM. When calculating the median or maximum likelihood, one essentially performs a non-linear operation on the solution set, and as a result, the median and maximum likelihoods are potentially invalid solutions to the LIM.
We now generalize the LA to over-determined LIM. To this end, we define an additional matrix equation that contains “approximate” equalities, which must be reproduced as closely as possible:
The form of A (“A” stands for approximate) and b is identical with E and f in equation (1), respectively, with coefficients as elements in matrix A that form linear equations with the flows in x and that have the corresponding numerical value in b. These “approximate” equalities are weighed with matrix S to assign the approximate equality with a respective uncertainty relative to one another. The scaling matrix S should be the variance–covariance matrix of the approximate equations, though in practice S is often a diagonal matrix with the observed variances (see for example, Menke 1984; Wunsch 1996). The approximate equation (7) causes the PDF to be no longer uniform: solutions that more closely fulfill the approximate equations have a higher probability than other solutions.
This is illustrated in a pedagogical model involving two flows and three approximate equality constraints (Figure 2A). Each approximate equality equation has an associated uncertainty, which is imposed through matrix S in equation (7), resulting for example from natural variability. This uncertainty is visualized by the grey shading bands around the equalities (Figure 2A). Hence, each equality (equation 3) defines a fixed line in the solution domain, whereas an approximate equality defines a region in the solution domain where the approximate equality is reproduced with certain likelihood. The superposition of likelihoods of the approximate equalities ultimately gives a complex likelihood field over the whole solution domain. Unlike for under-determined LIM, there is no black-and-white distinction between consistent and inconsistent solutions, but a more subtle distinction of more likely and less likely solutions. A representative set of solutions from such a solution domain is drawn with a Markov chain using, for example, the Metropolis algorithm (Van den Meersche and others 2009, schematically visualized in Figure 2A). The resulting solution set approximates the mPDF of each flow (Figure 2B).
Schematic representation of the LA for an over-determined LIM. Each of the three equality constraints is given a degree of uncertainty in (A) in which a darker shade of grey indicates a higher likelihood for the respective solution. The broader the uncertainty zone, the greater the uncertainty of the associated constraint. A sampling algorithm samples the solution domain (an arbitrary selection of sampled solutions is here indicated with the numbers 1–4), which leads to the PDF for each flow as shown in (B) for flow 1. See “Linear inverse modeling: three solution approaches”–“Likelihood approach” for more details.
An Example Application: C and N Cycling in Soils
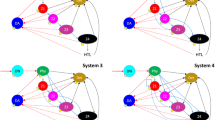
To illustrate the solution methods SSAunder, SSAover, and LA, we applied them to a simple soil food web. The food web topology and the associated parameters are taken from a recent study on the effects of fauna on C and N cycling in soils by Osler and Sonimerkorn (2007). The food web includes 5 compartments that are linked with 14 C and N flows (Figure 3). The complete data set consists of (high-quality) empirical data obtained from a study on a short-grass prairie food web (Hunt and others 1987), supplemented by (low-quality) literature data on soil food webs. As no ranges were given for the literature data, we introduced uncertainty by assuming that the reported value had a range of ±25% (this error value does not influence the conclusions drawn from the example food web). All scenarios were solved in the R-software (R Development Core Team 2008) using the newly developed packages ‘limSolve’ and ‘LIM’ (see Supplementary materials).
Topology of the example food web model. Five compartments are incorporated: particulate organic matter (POM), dissolved organic matter (DOM), microbes (MIC), fauna (FAU), and ammonium (NH4). The following C and N flows are incorporated: Plant litter deposits on the POM pool of the soil (f 1), POM dissolves to DOM (f 2), and DOM is taken up by microbes (f 3). Fauna consumes microbes (f 5) and POM (f 7) and the unassimilated food fraction flows to DOM (f 6). Fauna (f 10, f 9) and microbes (f 11, f 13) mineralize C to CO2 and N to NH4, but only microbes can take up NH4 (immobilization, f 12). Mortality of microbes (f 4) and fauna (f 8) is described as a flux to POM. Finally, plants take up N from the NH4 pool (f 14). Stocks are given in brackets as (C;N) in mmol m−2.
As a quality distinction is made between data sources, the site-specific data are implemented as “hard” equalities and literature data as “soft” inequalities, and so an under-determined LIM is obtained (Table 1). The LAunder solution of this LIM is presented in two ways (Figure 4): as histograms to reflect the mPDF of each flow and as paired plots to portray correlations between flow pairs. Note that the histograms are simply projections of the paired plots onto the axis of one of the flows as in Figure 1. An important advantage of the LAunder solution is that the probability distributions of all flows are revealed. The shapes of the mPDF differ between the flows: some flows have a bell-shaped distribution (for example, \( {\text{POM}}_{\text{C}} \to {\text{FAU}}_{\text{C}} ), \) other flows have a rather uniform distribution (for example, \( {\text{MIC}}_{\text{C}} \to {\text{POM}}_{\text{C}} ) \) and again others are strongly skewed toward their extremes (for example, \( {\text{MIC}}_{\text{C}} \to {\text{FAU}}_{\text{C}} ). \) The pair plots show correlations between flows, which can be given a direct ecological interpretation: for example, POM hydrolysis \( \left( {{\text{POM}}_{\text{C}} \to {\text{DOM}}_{\text{C}} } \right) \) correlates positively with microbial DOM uptake \( \left( {{\text{DOM}}_{\text{C}} \to {\text{MIC}}_{\text{C}} } \right) \) and respiration \( \left( {{\text{MIC}}_{\text{C}} \to {\text{DIC}}_{\text{C}} } \right). \) The SSAunder solution is indicated by the dashed line in the histograms. Note that this always lies at the extremes, with high flow magnitudes lying at the lower extreme and low flow magnitudes at the upper extreme (Figure 4). This “stickiness” of flow values to the extremes is known from the Kuhn-Tucker theorem that states that the solution is minimum only if one can make the solution more “simpler” or more “parsimonious,” that is, lower sum of squared flow values (equation 6), if one would cross some constraints (Menke 1984). This can also be understood intuitively, because the parsimony principle dictates that C goes as directly as possible where it is required according to the LIM and non-necessary C as determined by the LIM exits the system by the shortest possible route (Niquil and others 1998).
Histograms and pair plots of the C flows (mmol C m−2 d−1) of the example food web model calculated with LAunder. The x- and y-axes are scaled to the maximum flow values allowed within the LIM (that is, flow ranges). The dashed line in the histograms denotes the SSAunder solution and the solid line is the mean of the complete set of solutions. The flow DEP_C → POM_C is the deposition of plant litter. Note that the N flows are omitted to improve the readability of the figure. See “Linear inverse modeling: three solution approaches” for methodological details on the solution approaches SSAunder and LAunder.
The advantages of the LA are clear, it represents a complete solution of the LIM, because the distribution of all possible solutions is visualized and one can infer how well flows are constrained by the data. We extract a single “best” solution from the ensemble by averaging, which is, as argued above, a single solution that can be extracted from the solution set that is always consistent with the LIM equations. Moreover, the average flow values are very close to the medians (R 2 > 0.99, with a maximum relative deviation of 18% and absolute deviation of 1.3 mmol C m−2 d−1) and are central estimates of the flow values. The average flow values are represented in the histograms as solid lines (Figure 4).
When all available data are implemented as “hard” equalities (either in equation 3 or 7), one obtains an over-determined LIM (Table 1 and Supplementary materials). This LIM was solved with LAover and SSAover and the solutions are presented in Figure 5. The paired plots show a less uniform distribution over the solution domain as compared to LAunder, which is due to the fact that “fringe solutions” are less likely because their model–data fit is poorer. Hence, the center of the solution set truly gives a better fit to the imposed data as compared to the fringes. However, the correlations between flows as found in the under-determined LIM also surface here, for example, between microbial DOM uptake \( \left( {{\text{DOM}}_{\text{C}} \to {\text{MIC}}_{\text{C}} } \right) \) and respiration \( \left( {{\text{MIC}}_{\text{C}} \to {\text{DIC}}_{\text{C}} } \right). \) The histograms of all flows have a distinct maximum, which is very well approached by the SSAover, the latter being indicated by a dashed line in the histograms. The SSAover finds the solution that has an optimal model–data fit, which explains why it is very close to the maximum likelihoods of the flows in LAover. The SSAover thus represents a good and quick solution method when one is solely interested in the “best” estimate, the LAover however additionally gives information on the uncertainty associated with the estimation of a flow and correlations among the food web flows.
Histograms and pair plots of the C flows (mmol C m−2 d−1) of the example food web model calculated with LAover. The dashed line in the histograms denotes the SSAover solution. The flow DEP_C → POM_C is the deposition of plant litter. Note that the N flows are omitted from this figure to improve the readability of the figure. See “Linear inverse modeling: three solution approaches” for methodological details on the solution approaches SSAover and LAover.
Although this analysis cannot be regarded as a rigorous comparison of solution approaches, some important points surface: (1) The LA method provides much more information than the single-solution methods, because it reveals flow ranges, correlations between flows and probability distributions of the flows. (2) The optimization criterion that is used to select the SSAunder solution pushes the flow values to lower/upper extremes of their ranges. (3) The SSAover corresponds closely to the maximum likelihood of the LAover and represents a good and quick “best” estimate. (4) The only single valid solution that can be extracted from the solution set of LAunder is the average flow value, which represents a central estimate.
Discussion
Application of LIM in Food Web Research
Food web ecologists have realized in the recent years that the values of food web flows can differ over order of magnitudes, and that these differences are crucial for the functioning of food webs (Neutel and others 2002; Banašek-Richter and others 2004; Woodward and others 2005). We show here that linear inverse modeling (1) forms a promising tool for the quantification of real food webs in a systematic and standardized way, and that (2) the methodology is able to incorporate a variety of empirical data. We have also shown that the two existing approaches for solving such linear inverse food web models focus on a single solution of the model, whereas in reality an infinite number of solutions are equally likely from a “data-perspective.” Moreover, these single-solution approaches require additional assumptions to select a single solution. One approach involves placing lower-quality literature data from other food webs on equal footing to the data set from the food web under study (SSAover). The other approach distinguishes between high- and low-quality data, but requires a selection criterion to single out a solution (SSAunder). It remains to be investigated whether this selection criterion indeed selects the most likely food web structure.
Here we advance a LA, which has three important advantages: (1) data can be distinguished based on their quality, allowing data to be implemented as either “hard” equalities or “soft” inequalities, (2) both under- and over-determined LIM are solved using the same approach, and (3) rather than focusing on a single solution, the LA generates a marginal mPDF for each flow from which the distribution of flow values can be derived. The newly developed R-packages LIM and limSolve make the LA freely available (Supplementary materials show the setup and solution of the example model).
The complete set of food web structures generated with the LA can also be conveniently used in subsequent analysis of the food web. For example, Bersier and others (2002) developed food web descriptors, such as omnivory index and food chain length, which can be calculated from a quantitative food web. These calculations are typically inferred from a single food web structure. With the solution set generated by the LA, however, the food web descriptors can be calculated for every food web in the solution set. In this way, the distribution of flow values propagates to the distribution of the descriptors and one can assess how robust the food web descriptors are inferred (for example, Kones and others 2009). A similar strategy can be followed in the stability analysis of food webs (Neutel and others 2002; Rooney and others 2006), analysis of food quality on food web dynamics (Gaedke and others 2002) and modeling bioaccumulation of toxicants in food webs (De Laender and others 2009).
When implementing LIM, one important prerequisite is that all data are associated with appropriate temporal and spatial scales. For example, combining primary production rates measured over a period of hours with nutrient depletion rates derived from weekly monitoring is bound to give unreliable results. However, when the scaling issue is addressed, LIM can be used for the reconstruction of metabolic pathways in bacteria (Segre and others 2002), ocean circulation patterns (Wunsch 1996), and as we show here, food web flows. In terms of spatial scaling, food web applications typically consider the ecosystem as one homogeneous environment (as in the example model). However, this spatial homogeneity is no intrinsic limitation of LIM. Food webs can also be spatially resolved, for example, Breed and others (2004) modeled the planktonic food web of the Mississippi River plume in four coupled regions (~10–100 km scale). Jackson and Eldridge (1992) modeled C and N flows in two layers of the water column (~10–100 m scale).
Incorporating Novel Data Sources
The amount of data included in the LIM equalities determines whether the model is under- or over-determined (see above) and until now, we have discussed the implementation and solution of a food web LIM given some predefined data set. Although LAunder has clear advantages over single-solution approaches (as shown above), it also clearly illuminates some nagging problems of working with under-determined LIM: each valid solution of the under-determined LIM is equally likely (Figure 1A) and as a result the mPDF of many flows have a comparatively uniform distribution and are thus not very well constrained (Figure 4). This is different for the over-determined LIM, where a true maximum-likelihood solution exists (Figure 2A) and where many of the mPDF of the flows are bell-shaped with distinct maxima (Figure 5). A proactive response of the food web researcher would be to try to include as much high-quality data as equalities as possible, such that the food web LIM will finally become over-determined. This will be difficult to achieve with the data types that are currently exploited, because these typically constrain only how much food is required, but provide limited information from where this food is obtained. We therefore end this article with suggestions of a variety of data types that can be straightforwardly implemented in food web LIM, but that are currently not or only limitedly used. Their implementation will decrease the degree of under-sampling of food webs, reduce the uncertainty of the estimated flow values and will bring over-determined food web models within reach.
Elemental stoichiometry couples empirical data on flows in one currency to flows in another currency (for example, Vézina and Platt 1988; Jackson and Eldridge 1992; Gaedke and others 2002). In the example model, grazing on microbial C and N is coupled through the bacterial C:N ratio (see Table 1). Stoichiometric relations have not been used to make food web processes dependent on the observed elemental compositions of the compartments. Experiments show that the gross growth efficiency decreases when the elemental imbalance between predator and prey increases (chapter 5 in Sterner and Elser 2002). Similar stoichiometric relations have been reported between the C:P ratio of autotrophs and their specific growth rate (chapter 3 in Sterner and Elser 2002) and between the elemental composition of detritus and its decomposition rate (Cebrian 2004). This information can be used by making the value of the associated parameters (for example, gross growth efficiency, specific growth rate, decomposition rate) directly dependent on the observed stoichiometric compositions. When such relations are implemented, they will lead to narrower and more realistic lower and/or upper values in the matrix inequalities equation and thus better constrained flow values.
Stable isotope signatures provide time-integrated information on the trophic level of an organism (Minagawa and Wada 1984; Post 2002) and its diet composition (Phillips and Gregg 2003) under natural conditions. This knowledge cannot be derived from traditional data on C or N processing and therefore stable isotope signatures are now a standard method in food web research. The standard method to interpret stable isotope data is by means of a linear mixing model and these can be seamlessly integrated in a LIM. It is therefore surprising that stable isotope data have not been frequently used in food web LIM. The great advantage is that they exclude diet combinations that are in conflict with the isotope data. This exclusion strongly narrows the flow ranges in the food web. A clear illustration is provided by a recent study of an estuarine intertidal food web, where δ13C data were combined with conventional data on C cycling (Van Oevelen and others 2006). The δ13C data distinguished diet contributions from benthic microalgae (heavy δ13C) from those of phytoplankton and detritus (lighter δ13C). The uncertainty range of many flows decreased significantly after the addition of stable isotope data to the LIM. A similar observation was made in a LIM study on the structure of the pelagic food web in an estuary, where the incorporation of δ13C data greatly influenced the heterotrophic flows in the food web LIM (Eldridge and others 2005).
Another method to infer the diet of an organism is based on its fatty acid composition. In general, the fatty acid composition of an organism reflects that of its resource, barring some alterations during digestion and deposition. Fatty acid signatures have been used to establish the presence or absence of feeding links between compartments (for example, Meziane and Tsuchiya 2000). Recently, Iverson and others (2004) provided a quantitative approach to reconstruct diet compositions from fatty acid signatures. The model assumes linear mixing of fatty acid compositions and is thus analogous to the mixing model of stable isotopes. The model can account for biosynthetic alterations by means of fatty acid-specific calibration coefficients and specification of the total lipid content of the prey (Iverson and others 2004). In theory, fatty acid compositions could significantly extend the empirical data set of a LIM and thus aid in resolving large and complex food webs.
In conclusion, we have shown that LIM are powerful tools to quantitatively reconstruct food webs by merging a variety of traditional data types (for example, biomass) and currently under-exploited data sources (for example, stable isotopes). The LA proposed here solves a LIM without additional assumptions, and robustly calculates the “best” flow value, its uncertainty and correlations with other flows. In this way, LIM provide an effective tool to bridge the gap between incomplete and uncertain empirical data on natural food webs and the analysis of food web structures.
References
Banašek-Richter C, Cattin MF, Bersier LF. 2004. Sampling effects and the robustness of quantitative and qualitative food-web descriptors. J Theor Biol 226:23–32.
Berlow EL, Neutel AM, Cohen JE, De Ruiter PC, Ebenman B, Emmerson M, Fox JW, Jansen VAA, Jones JI, Kokkoris GD, Logofet DO, McKane AJ, Montoya JM, Petchey O. 2004. Interaction strengths in food webs: issues and opportunities. J Anim Ecol 73:585–98.
Bersier LF, Banasek-Richter C, Cattin MF. 2002. Quantitative descriptors of food-web matrices. Ecology 83:2394–407.
Breed GA, Jackson GA, Richardson TL. 2004. Sedimentation, carbon export and food web structure in the Mississippi River plume described by inverse analysis. Mar Ecol Prog Ser 278:35–51.
Cebrian J. 2004. Patterns of herbivory and decomposition in aquatic and terrestrial ecosystems. Ecol Monogr 74:237–59.
Christensen V, Pauly D. 1992. Ecopath-II—a software for balancing steady-state ecosystem models and calculating network characteristics. Ecol Modell 61:169–85.
Conover RJ. 1966. Factors affecting the assimilation of organic matter by zooplankton and the question of superfluous feeding. Limnol Oceanogr 11:346–54.
De Laender F, Van Oevelen D, Soetaert K, Middelburg JJ. 2009. Incorporating ecological data and associated uncertainty in bioaccumulation modeling: methodology development and case study. Environ Sci Technol 43:2620–6.
De Ruiter PC, Neutel AM, Moore JC. 1995. Energetics, patterns of interaction strengths, and stability in real ecosystems. Science 269:1257–60.
Diffendorfer JE, Richards PM, Dalrymple GH, DeAngelis DL. 2001. Applying linear programming to estimate fluxes in ecosystems or food webs: an example from the herpetological assemblage of the freshwater Everglades. Ecol Modell 144:99–120.
Eldridge PM, Cifuentes LA, Kaldy JE. 2005. Development of a stable isotope constraint system for estuarine food web models. Mar Ecol Prog Ser 303:73–90.
Gaedke U, Hochstadter S, Straile D. 2002. Interplay between energy limitation and nutritional deficiency: empirical data and food web models. Ecol Monogr 72:251–70.
Hunt HW, Coleman DC, Ingham ER, Ingham RE, Elliott ET, Moore JC, Rose SL, Reid CPP, Morley CR. 1987. The detrital food web in a shortgrass prairie. Biol Fertil Soils 3:57–68.
Iverson SJ, Field C, Bowen WD, Blanchard W. 2004. Quantitative fatty acid signature analysis: a new method of estimating predator diets. Ecol Monogr 74:211–35.
Jackson GA, Eldridge PM. 1992. Food web analysis of a planktonic system off Southern California. Prog Oceanogr 30:223–51.
Klepper O, Van de Kamer JPG. 1987. The use of mass balances to test and improve the estimates of carbon fluxes in an ecosystem. Math Biosci 85:37–49.
Kones JK, Soetaert K, van Oevelen D, Owino JO. 2009. Are network indices robust indicators of food web functioning? A Monte Carlo approach. Ecol Modell 220:370–82.
Kones JK, Soetaert K, van Oevelen D, Owino JO, Mavuti K. 2006. Gaining insight into food webs reconstructed by the inverse method. J Mar Syst 60:153–66.
Menke W. 1984. Geophysical data analysis: discrete inverse theory. New York: Academic Press.
Meziane T, Tsuchiya M. 2000. Fatty acids as tracers of organic matter in the sediments and food web of a mangrove/intertidal flat ecosystem, Okinawa. Japan Mar Ecol Prog Ser 200:49–57.
Minagawa M, Wada E. 1984. Stepwise enrichment of 15N along food chains: further evidence and the relation between δ15N and animal age. Geochim Cosmochim Acta 48:1135–40.
Neutel AM, Heesterbeek JAP, de Ruiter PC. 2002. Stability in real food webs: weak links in long loops. Science 296:1120–3.
Niquil N, Jackson GA, Legendre L, Delesalle B. 1998. Inverse model analysis of the planktonic food web of Takapoto Atoll (French Polynesia). Mar Ecol Prog Ser 165:17–29.
Osler GHR, Sonimerkorn M. 2007. Toward a complete soil C and N cycle: incorporating the soil fauna. Ecology 88:1611–21.
Phillips DL, Gregg JW. 2003. Source partitioning using stable isotopes: coping with too many sources. Oecologia 136:261–9.
Pimm SL, Lawton JH, Cohen JE. 1991. Food web patterns and their consequences. Nature 350:669–74.
Post DM. 2002. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83:703–18.
R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. http://www.R-project.org.
Richardson TL, Jackson GA, Burd AB. 2003. Planktonic food web dynamics in two contrasting regions of Florida Bay, USA. Bull Mar Sci 73:569–91.
Rooney N, McCann K, Gellner G, Moore JC. 2006. Structural asymmetry and the stability of diverse food webs. Nature 442:265–9.
Segre D, Vitkup D, Church GM. 2002. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci 99:15112–17.
Soetaert K, Van den Meersche K, Van Oevelen D. 2008. limSolve: solving linear inverse models. R package version 1.5.1. http://lib.stat.cmu.edu/R/CRAN/web/packages/limSolve/index.html.
Soetaert K, Van Oevelen D. 2008. LIM: linear inverse model examples and solution methods. R package version 1.4. http://lib.stat.cmu.edu/R/CRAN/web/packages/LIM/index.html.
Steele JH. 2009. Assessment of some linear food web methods. J Mar Syst 76:186–94.
Sterner RW, Elser J. 2002. Ecological stoichiometry. The biology of elements from molecules to the biosphere. Princeton: Princeton University Press.
Van den Meersche K, Soetaert K, Van Oevelen D. 2009. xsample(): an R function for sampling linear inverse problems. J Stat Softw 30:1–15.
Van Oevelen D, Soetaert K, Middelburg JJ, Herman PMJ, Moodley L, Hamels I, Moens T, Heip CHR. 2006. Carbon flows through a benthic food web: integrating biomass, isotope and tracer data. J Mar Res 64:1–30.
Vézina AF, Berreville F, Loza S. 2004. Inverse reconstructions of ecosystem flows in investigating regime shifts: impact of the choice of objective function. Prog Oceanogr 60:321–41.
Vézina AF, Platt T. 1988. Food web dynamics in the ocean. I. Best-estimates of flow networks using inverse methods. Mar Ecol Prog Ser 42:269–87.
Williams RJ, Martinez ND. 2000. Simple rules yield complex food webs. Nature 404:180–3.
Woodward G, Speirs DC, Hildrew AG. 2005. Quantification and resolution of a complex, size-structured food web. Adv Ecol Res 36:85–135.
Wunsch C. 1996. The ocean circulation inverse problem. New York: Cambridge University Press.
Acknowledgements
Peter Herman, Carlo Heip, Matthijs Vos, Johan van de Koppel, and two anonymous reviewers are thanked for constructive feedback. This research was supported by the HERMES project, EC Contract no. GOCE-CT-2005-511234, funded by the European Commission’s Sixth Framework Program under the priority ‘Sustainable Development, Global Change and Ecosystems’ and the Netherlands Organisation of Scientific Research (PIONIER 833.02.2002). K.V.d.M. was supported by a grant from the Flemish Fund for Scientific Research (FWO), and extra funding from the University of Gent. This is publication 4656 of the Netherlands Institute of Ecology (NIOO-KNAW), Yerseke.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author Contributions
Dick van Oevelen, Karline Soetaert, and Jack J. Middelburg have conceived of or designed the study; Dick van Oevelen and Karline Soetaert performed research; Dick van Oevelen and Karline Soetaert analyzed data; Dick van Oevelen, Karline Soetaert, and Karel Van den Meersche have contributed new methods or models; Dick van Oevelen, Karline Soetaert, Jack J. Middelburg, Filip J. R. Meysman, and Alain F. Vézina have written the article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Oevelen, D., Van den Meersche, K., Meysman, F.J.R. et al. Quantifying Food Web Flows Using Linear Inverse Models. Ecosystems 13, 32–45 (2010). https://doi.org/10.1007/s10021-009-9297-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10021-009-9297-6