Abstract
We have developed a fully automated method, InterProSurf, to predict interacting amino acid residues on protein surfaces of monomeric 3D structures. Potential interacting residues are predicted based on solvent accessible surface areas, a new scale for interface propensities, and a cluster algorithm to locate surface exposed areas with high interface propensities. Previous studies have shown the importance of hydrophobic residues and specific charge distribution as characteristics for interfaces. Here we show differences in interface and surface regions of all physical chemical properties of residues as represented by five quantitative descriptors. In the current study a set of 72 protein complexes with known 3D structures were analyzed to obtain interface propensities of residues, and to find differences in the distribution of five quantitative descriptors for amino acid residues. We also investigated spatial pair correlations of solvent accessible residues in interface and surface areas, and compared log-odds ratios for interface and surface areas. A new scoring method to predict potential functional sites on the protein surface was developed and tested for a new dataset of 21 protein complexes, which were not included in the original training dataset. Empirically we found that the algorithm achieves a good balance in the accuracy of precision and sensitivity by selecting the top eight highest scoring clusters as interface regions. The performance of the method is illustrated for a dimeric ATPase of the hyperthermophile, Methanococcus jannaschii, and the capsid protein of Human Hepatitis B virus. An automated version of the method can be accessed from our web server at http://curie.utmb.edu/prosurf.html.

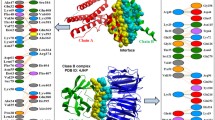
Comparison of the experimentally observed interface (blue) of the dimeric ATPase of the hyperthermophile Methanococcus jannaschii with the prediction of InterProSurf using a cluster (red) and patch (yellow) algorithm.








Similar content being viewed by others
References
Kinoshita K, Nakamura H (2003) Curr Opin Struct Biol 13:396–400
Archakov AI, Govorun VM, Dubanov AV, Ivanov YD, Veselovsky AV, Lewi P, Janssen P (2003) Proteomics 3:380–391
Lo Conte L, Chothia C, Janin J (1998) J Mol Biol 285:2177–2198
Janin J, Chothia C (1990) J Bio Chem 265:16027–16030
Tsai CJ, Lin SL, Wolfson HJ, Nussinov R (1997) Protein Sci 6:53–64
Hu ZJ, Ma BY, Wolfson H, Nussinov R (2000) Proteins-Structure Function Genetics 39:331–342
Jones S, Thornton JM (1996) Proc Natl Acad Sci USA 93:13–20
McCoy AJ, Epa VC, Colman PM (1997) J Mol Biol 268:570–584
Thorn KS, Bogan AA (2001) Bioinformatics 17:284–285
Bader GD, Betel D, Hogue CWV (2003) Nucleic Acids Res 31:248–250
Xenarios I, Salwinski L, Duan XQJ, Higney P, Kim SM, Eisenberg D (2002) Nucleic Acids Res 30:303–305
Xenarios L, Eisenberg D (2001) Curr Opin Biotechnol 12:334–339
DeLano WL (2002) Curr Opin Struct Biol 12:14–20
DeLano WL, Ultsch MH, de Vos AM, Wells JA (2000) Science 287:1279–1283
Kortemme T, Baker D (2002) Proc Natl Acad Sci USA 99:14116–14121
Bogan AA, Thorn KS (1998) J Mol Biol 280:1–9
Clackson T, Wells JA (1995) Science 267:383–386
Jones S, Thornton JM (1997) J Mol Biol 272:133–143
Jones S, Thornton JM (1997) J Mol Biol 272:121–132
Jones S, Thornton JM (1995) Prog Biophys Mol Biol 63:31–35
Jones S, Marin A, Thornton JM (2000) Protein Eng 13:77–82
Bordner AJ, Abagyan R (2005) Proteins: Structure, Function, Bioinformatics 60:353–366
Murakami Y, Jones S (2006) Bioinformatics 22:1794–1795
Brinda KV, Kannan N, Vishveshwara S (2002) Protein Eng 15:265–277
Landgraf R, Xenarios I, Eisenberg D (2001) J Mol Biol 307:1487–1502
Morrison KL, Weiss GA (2001) Curr Opin Chem Biol 5:302–307
Kortemme T, Kim DE, Baker D (2004) Sci STKE 219:12
Massova I, Kollman PA (1999) J Am Chem Soc 121:8133–8143
Zhou HX, Shan Y (2001) Proteins 44:336–343
Fariselli P, Pazos F, Valencia A, Casadio R (2002) Eur J Biochem 269:1356–1361
Koike A, Takagi T (2004) Protein Engineering Design & Selection 17:165–173
Bradford JR, Westhead DR (2004) Bioinformatics 21:1487–1494
Gallet X, Charloteaux B, Thomas A, Brasseur R (2000) J Mol Biol 302:917–926
Bock JR, Gough DA (2001) Bioinformatics 17:455–460
Gao Y, Wang RX, Lai LH (2004) J Mol Model 10:44–54
Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, Kavraki L, Lichtarge O (2003) J Mol Biol 326:255–261
Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N (2003) Bioinformatics 19:163–164
Chakrabarti P, Janin J (2002) Proteins 47:334–343
Bahadur RP, Chakrabarti P, Rodier F, Janin J (2004) J Mol Biol 336:943–955
Glaser F, Steinberg DM, Vakser IA, Ben-Tal N (2001) Proteins-Structure Function Genetics 43:89–102
Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N (2005) Nucleic Acids Res 33:W299–W302
Neuvirth H, Raz R, Schreiber G (2004) J Mol Biol 338:181–199
Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A (2005) Nucleic Acids Res 33:W331–W336
Mihalek I, Res I, Lichtarge O (2006) Bioinformatics 22:1656–1657
Venkatarajan MS, Braun W (2001) J Mol Model 7:445–453
Negi SS, Kolokoltsov AA, Schein CH, Davey RA, Braun W (2006) J Mol Model 12:921–929
Altschul S, Madden T, Schaffer A, Zhang JH, Zhang Z, Miller W, Lipman D (1998) Faseb J 12:A1326–A1326
Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acids Res 22:4673–4680
Fraczkiewicz R, Braun W (1998) J Comp Chem 19:319
Liang S, Liu Z, Li W, Ni L, Lai L (2000) Biopolymers 54:515–523
Singh RK, Tropsha A, Vaisman II (1996) J Comp Biol 3:213–221
Linde Y, Buzo A, Gray RM (1980) IEEE Trans Commun 28:84–95
Sayood K (2000) Introduction to data compression, 2nd edn. Morgan Kaufmann Publishers Inc
Patane G, Russo M (2001) Neural Netw 14:1219–1237
Equitz HW (1989) IEEE Trans Acoustics Speech Signal Process 37:1568–1575
Cosman PC, Oehler KL, Riskin EA, Gray RM (1993) Proc I E E E 81:1326–1341
Lin CL, Tai SC (1998) IEEE Trans Circuits Syst, II Analog Digit Signal Process 45:432–435
Baldi P, Brunak S, Chauvin Y, Andersen CAF, Nielsen H (2000) Bioinformatics 16:412–424
Ma B, Elkayam T, Wolfson H, Nussinov R (2003) Proc Natl Acad Sci USA 100:5772–5777
Nagano N, Ota M, Nishikawa K (1999) FEBS Lett 458:69–71
Karlin S, Zhu ZY, Baud F (1999) Proc Natl Acad Sci USA 96:12500–12505
Mathura VS, Schein CH, Braun W (2003) Bioinformatics 19:1381–1390
Zarembinski TI, Hung LW, Mueller-Dieckmann HJ, Kim KK, Yokota H, Kim R, Kim SH (1998) Proc Natl Acad Sci USA 95:15189–15193
Wynne SA, Crowther RA, Leslie AGW (1999) Mol Cell 3:771–780
Acknowledgements
This work was supported by National Institutes of Health Grants R21 AI055746 and R01 AI064913.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Negi, S.S., Braun, W. Statistical analysis of physical-chemical properties and prediction of protein-protein interfaces. J Mol Model 13, 1157–1167 (2007). https://doi.org/10.1007/s00894-007-0237-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-007-0237-0