Abstract
Alcohol dehydrogenase (ADH; EC: 1.1.1.1) is a key enzyme in production and utilization of ethanol. In this study, the gene encoding for ADH of the haloalkaliphilic archaeon Natronomonas pharaonis (NpADH), which has a 1,068-bp open reading frame that encodes a protein of 355 amino acids, was cloned into the pET28b vector and was expressed in Escherichia coli. Then, NpADH was purified by Ni-NTA affinity chromatography. The recombinant enzyme showed a molecular mass of 41.3 kDa by SDS-PAGE. The enzyme was haloalkaliphilic and thermophilic, being most active at 5 M NaCl or 4 M KCl and 70°C, respectively. The optimal pH was 9.0. Zn2+ significantly inhibited activity. The K m value for acetaldehyde was higher than that for ethanol. It was concluded that the physiological role of this enzyme is likely the catalysis of the oxidation of ethanol to acetaldehyde.



Similar content being viewed by others
References
Bieger B, Essen LO, Oesterhelt D (2003) Crystal structure of halophilic dodecin. A novel, dodecameric flavin binding protein from Halobacterium salinarum. Structure 11:375–385
Bonete MJ, Camacho ML, Cadenas E (1986) Purification and properties of NAD+-dependent glutamate dehydrogenase from Halobacterium halobium. Int J Biochem 18:785–789
Bonete MJ, Camacho ML, Cadenas E (1987) A new glutamate dehydrogenase from Halobacterium halobium with different coenzyme specificity. Int J Biochem 19:1149–1155
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bräsen C, Schönheit P (2004) Regulation of acetate and acetyl-CoA converting enzymes during growth on acetate and/or glucose in the halophilic archaeon Haloarcula marismortui. FEMS Microbiol Lett 241:21–26
Ceccarelli C, Liang ZX, Strickler M, Prehna G, Goldstein BM, Klinman JP, Bahnson BJ (2004) Crystal structure and amide H/D exchange of binary complexes of alcohol dehydrogenase from Bacillus stearothermophilus: Insight into thermostability and cofactor binding. Biochemistry 43:5266–5277
Danson MJ, Hough DW (1997) The structural basis of protein halophilicity. Comp Biochem Physiol 117A:307–312
Demirjian DC, Moris-Varas F, Cassidy CS (2001) Enzymes from extremophiles. Curr Opin Chem Biol 5:144–151
Dym O, Mevarech M, Sussman JL (1995) Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science 267:1344–1346
Ebel C, Costenaro L, Pascu M, Faou P, Kernal B, Proust-De Martin F, Zaccai G (2002) Solvent interactions of halophilic malate dehydrogenase. Biochemistry 41:13234–13244
Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D (2005) Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res 15:1336–1343
Forrest GL, Gonzalez B (2000) Carbonyl reductases. Chem Biol Interact 129: 21–40
Frolow F, Harel M, Sussman JL, Mevarech M, Shoham M (1996) Protein adaptation to a saturated salt environment: insights from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nature Struct Biol 3:451–457
Fukuchi S, Yoshimune K, Wakayama M, Moriguchi M, Nishikawa K (2003) Unique amino acid composition of proteins in halophilic bacteria. J Mol Biol 327:347–357
Irimia A, Ebel C, Madern D, Richard SB, Cosenza LW, Zaccai G, Vellieux FMD (2003) The oligomeric states of Haloarcula marismortui malate dehydrogenase are modulated by solvent components as shown by crystallographic and biochemical studies. J Mol Biol 326:859–873
Jörnvall H, Persson B, Jeffery J (1987) Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem 167:195–201
Kamekura M (1998) Diversity of extremely halophilic bacteria. Extremophiles 2:289–295
Konstantinidis K, Tebbe A, Klein C, Scheffer B, Aivaliotis M, Bisle B, Falb M, Pfeiffer F, Siedler F, Oesterhelt D (2007) Genome-wide proteomics of Natronomonas pharaonis. J Proteome Res 6:185–193
Kumar S, Tamura K, Nei M (1994) MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191
Kushner DJ (1978) Life in high salt and solute concentrations. In: Kushner DJ (ed) Microbial life in extreme environments. Academic Press, London, pp 317–368
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4:91–98
Madern D, Camacho M, Rodriguez-Arnedo A, Bonete MJ, Zaccai G (2004) Salt-dependent studies of NADP-dependent isocitrate dehydrogenase from the halophilic archaeon Haloferax volcanii. Extremophiles 8:377–384
Margesin R, Schinner F (2001) Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73–83
Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschröder M, Spudich JL, Jung KW, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S (2000) Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA 97:12176–12181
Oren A (1973) A thermophilic amyloglucosidase from Halobacterium sodomense, a halophilic bacterium from the Dead Sea. Curr Microbiol 8: 225–230
Padan E, Zilberstein D, Schuldiner S (1981) pH homeostasis in bacteria. Biochim Biophys Acta 650:151–166
Padan E, Bibi E, Ito M, Krulwich TA (2005) Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta 1717:67–88
Pieper U, Kapadia G, Mevarech M, Herzberg O (1998) Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure 6:75–88
Reid MF, Fewson CA (1994) Molecular characterization of microbial alcohol dehydrogenase. Crit Rev Microbiol 20:13–56
Richard SB, Madern D, Garcin E, Zaccai G (2000) Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 Å resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui. Biochemistry 39:992–1000
Saitou N, Nei M (1987) The neighbour joining method: a new tool for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
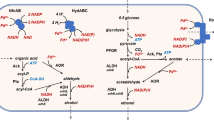
Siebers B, Schönheit P (2005) Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr Opin Microbiol 8:695–705
Yamada Y, Fujiwara T, Sato T, Igarashi N, Tanaka N (2002) The 2.0 Å crystal structure of catalase-peroxidase from Haloarcula marismortui. Nat Struct Biol 9:691–695
Ziegenhorn J, Senn M, Bücher T (1976) Molar absorptivities of β-NADH and β-NADPH. Clin Chem 22:151–160
Acknowledgments
This work was supported by the grants from the Major State Basic Research Development Program of China (973 Program) (Grant No. 2004CB719604-3) and the National Natural Science Foundation of China (Grant No. 30670048).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi.
Rights and permissions
About this article
Cite this article
Cao, Y., Liao, L., Xu, Xw. et al. Characterization of alcohol dehydrogenase from the haloalkaliphilic archaeon Natronomonas pharaonis . Extremophiles 12, 471–476 (2008). https://doi.org/10.1007/s00792-007-0133-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-007-0133-7