Abstract
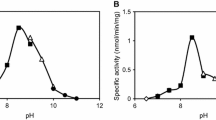
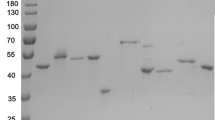
We expressed a histidine racemase from Leuconostoc mesenteroides subsp. sake NBRC 102480 (Lm-HisR) successively in a soluble fraction of Escherichia coli BL21 (DE3) and then highly purified it from the cell-free extract. Lm-HisR showed amino acid racemase activity on histidine specifically. This is the first example of an amino acid racemase specifically acting on histidine. Phylogenetic analysis of Lm-HisR showed that Lm-HisR was located far from the cluster of alanine racemases reported thus far and only in lactic acid bacteria of the genus Leuconostoc. Alignment of the primary structure of Lm-HisR with those of lysine and alanine racemases and alanine racemase homologs previously reported revealed that the PLP-binding lysine and catalytic tyrosine were completely conserved, and some residues that are unique to the phylogenetic branch of Lm-HisR, Phe44, Ser45, Thr174, Thr206, His286, Ser287, Phe292, Gly312, Val357, and Ala358 were identified. We determined the crystal structure of Lm-HisR complexed with PLP at a 2.1-Å resolution. The crystal structure contained four molecules (two dimers) in the asymmetric unit. When comparing the 3D structure of Lm-HisR with those of racemases from Geobacillus stearothermophilus and Oenococcus oeni, Met315 was completely conserved, but Val357 was not. In addition, two significant differences were observed between Lm-HisR and G. stearothermophilus alanine racemase. Phe44 and His286 in Lm-HisR corresponded to Tyr43 and Tyr284 in G. stearothermophilus alanine racemase, respectively. Based on the structural analysis, comparison with alanine racemase, and docking simulation, three significant residues, Phe44, His286, and Val357, were identified that may control the substrate specificity of Lm-HisR.




Similar content being viewed by others
Abbreviations
- ASA:
-
Accessible surface area
- Lm-HisR:
-
Histidine racemase from Leuconostoc mesenteroides subsp. sake NBRC 102480
- PDB:
-
Protein data bank
- PLP:
-
Pyridoxal 5´-phosphate
- RMSD:
-
Root mean square deviations
References
Adams E, Norton IL (1964) Purificatio and properties of inducible hydroxyproline 2-epimerase from Pseudomonas. J Biol Chem 239:1525–1535
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66:213–221
Briggs GE, Haldane JB (1925) A note on the kinetics of enzyme action. Biochem J 19:338–339
Chen HP, Lin CF, Lee YJ, Tsay SS, Wu SH (2000) Purification and properties of ornithine racemase from Clostridium sticklandii. J Bacteriol 182:2052–2054
Espaillat A, Carrasco-López C, Bernardo-García N, Pietrosemoli N, Otero LH, Álvarez L, de Pedro MA, Pazos F, Davis BM, Waldor MK, Hermoso JÁ, Cava F (2014) Structural basis for the broad specificity of a new family of amino-acid racemases. Acta Cryst D 70:79–90 ci
Gogami Y, Ito K, Kamitani Y, Matsushima Y, Oikawa T (2009) Occurrence of d-serine in rice and characterization of rice serine racemase. Phytochemistry 70:380–387
Gogami Y, Okada K, Oikawa T (2011) High-performance liquid chromatography analysis of naturally occurring d-amino acids in sake. J Chromatogr B Analyt Technol Biomed Life Sci 879:3259–3267
Gogami Y, Okada K, Oikawa T (2012) Quantitative analysis of d-amino acids in sake brewing processes of Kimoto, Kimoto adding starter latctic acid bacteria, and Sokujomoto. Trace Nutr Res 29:1–6
Grishin NV, Phillips MA, Goldsmith EJ (1995) Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci 4:1291–1304
Hoffman HE, Jirásková J, Ingr M, Zvelebil M, Konvalinka J (2009) Recombinant human serine racemase: enzymologic characterization and comparison with its mouse ortholog. Protein Expr Purif 63:62–67
Hor L, Dobson RC, Downton MT, Wagner J, Hutton CA, Perugini MA (2013) Dimerization of bacterial diaminopimelate epimerase is essential for catalysis. J Biol Chem 288:9238–9248
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Kato S, Oikawa T (2018) A novel bifunctional amino acid racemase with multiple substrate specificity, MalY from Lactobacillus sakei LT-13: genome-based identification and enzymological characterization. Front Microbiol 9:403
Kato S, Hemmi H, Yoshimura T (2012) Lysine racemase from a lactic acid bacterium, Oenococcus oeni: structural basis of substrate specificity. J Biochem 152:505–508
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK (2009) d-Amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:552–555
Long Z, Lee JA, Okamoto T, Sekine M, Nimura N, Imai K, Yohda M, Maruyama T, Sumi M, Kamo N, Yamagishi A, Oshima T, Homma H (2001) Occurrence of d-amino acids and a pyridoxal 5′-phosphate-dependent aspartate racemase in the acidothermophilic archaeon, Thermoplasma acidophilum. Biochem Biophys Res Commun 281(2):317–321
Long F, Vagin AA, Young P, Murshudov GN (2008) BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr 64:125–132
Matsui D, Oikawa T (2010) Detection and function of the intramolecular disulfide bond in arginine racemase: an enzyme with broad substrate specificity. Chem Biodivers 7:591–602
Matsui D, Oikawa T, Arakawa N, Osumi S, Lausberg F, Stäbler N, Freudl R, Eggeling L (2009) A periplasmic, pyridoxal-5′-phosphate-dependent amino acid racemase in Pseudomonas taetrolens. Appl Microbiol Biotechnol 83:1045–1054
McRee DE (1992) A visual protein crystallographic software system for X11/XView. J Mol Graph 10:44–46
Miyamoto T, Katane M, Saitoh Y, Sekine M, Homma H (2017) Identification and characterization of novel broad-spectrum amino acid racemases from Escherichia coli and Bacillus subtilis. Amino Acids 49:1885–1894
Miyamoto T, Katane M, Saitoh Y, Sekine M, Homma H (2018) Cystathionine β-lyase is involved in d-amino acid metabolism. Biochem J 475:1397–1410
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 16:2785–2791
Oikawa T, Tauch A, Schaffer S, Fujioka T (2006) Expression of alr gene from Corynebacterium glutamicum ATCC 13032 in Escherichia coli and molecular characterization of the recombinant alanine racemase. J Biotechnol 125:503–512
Okada K, Gogami Y, Oikawa T (2013) Principal component analysis of the relationship between the d-amino acid concentrations and the taste of the sake. Amino Acids 44:489–498
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326
Palani K, Burley SK, Swaminathan S (2013) Structure of alanine racemase from Oenococcus oeni with bound pyridoxal 5′-phosphate. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:5–9
Radkov AD, Moe LA (2013) Amino acid racemase in Pseudomonas putida KT2440. J Bacteriol 195:5016–5024
Rahamanian M, Waller GR, Grady Smith W (1971) Biosynthesis of d-aspartic acid by Streptococcus faecalis. J Biol Chem 246:823–830
Rudnick G, Abeles RH (1975) Reaction mechanism and structure of the active site of proline racemase. Biochemistry 14:4515–4522
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shaw JP, Petsko GA, Ringe D (1997) Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-Å resolution. Biochemistry 36:1329–1342
Soda K, Osumi T (1969) Crystalline amino acid racemase with low substrate specificity. Biochem Biophys Res Commun 35:363–368
Stein T, Kluge B, Vater J, Franke P, Otto A, Wittmann-Liebold B (1995) Gramicidin S synthetase 1 (phenylalanine racemase), a prototype of amino acid racemases containing the cofactor 4′-phosphopantetheine. Biochemistry 34:4633–4642
Sun X, He G, Wang X, Xu S, Ju J, Xu X (2015) Crystal structure of a thermostable alanine racemase from Thermoanaerobacter tengcongensis MB4 reveals the role of Gln360 in substrate selection. PLoS One 2810:e0133516
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Uo T, Yoshimura T, Shimizu S, Esaki N (1998) Occurrence of pyridoxal 5′-phosphate-dependent serine racemase in silkworm, Bombyx mori. Biochem Biophys Res Commun 246:31–34
Watanabe T, Shibata K, Kera Y, Yamada R (1998) Occurrence of free d-aspartate and aspartate racemase in the blood shell Scapharca broughtonii. Amino Acids 14:353–360
Watanabe A, Yoshimura T, Mikami B, Hayashi H, Kagamiyama H, Esaki N (2002) Reaction mechanism of alanine racemase from Bacillus stearothermophilus: X-ray crystallographic studies of the enzyme bound with N-(5′-phosphopyridoxyl)alanine. J Biol Chem 277:19166–19172
Wild J, Hennig J, Lobocka M, Walczak W, Kłopotowski T (1985) Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet 198:315–322
Wu D, Hu T, Zhang L, Chen J, Du J, Ding J, Jiang H, Shen X (2008) Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis. Protein Sci 17(6):1066–1076
Yamaguchi T, Choi SY, Okada H, Yohda M, Kumagai H, Esaki N, Soda K (1992) Properties of aspartate racemase, a pyridoxal 5′-phosphate-independent amino acid racemase. J Biol Chem 267(26):18361–18364
Yoshimura T, Ashiuchi M, Esaki N, Kobatake C, Choi SY, Soda K (1993) Expression of glr (murI, dga) gene encoding glutamate racemase in Escherichia coli. J Biol Chem 268:24242–24246
Acknowledgements
The synchrotron radiation experiments were performed on the BL5A beamline at Photon Factory (proposal no. 2014G645). This work was also supported by JSPS KAKENHI grant number JP16KT0063 (awarded to MA and TO). This study was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Support Program for the Strategic Research Foundation at Private Universities (2013–2017), and the Kansai University Organization for Research and Development of Innovative Science and Technology (ORDIST) Grant (2018). We thank Dr. Yoshitaka Gogami for the construction of pET21b-Lm-hisR and characterization of the basic properties of Lm-HisR. Finally, we appreciate the contribution of Dr. Ryota Kuroki to promote in the early stages of this study.
Author information
Authors and Affiliations
Contributions
TO designed the studies. TO, MA, and SK wrote the paper. RS and SK conducted the experiments. TO and SK conducted the phylogenetic analysis. MA analyzed the X-ray crystal structure.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: C. Schiene-Fischer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adachi, M., Shimizu, R., Kato, S. et al. The first identification and characterization of a histidine-specific amino acid racemase, histidine racemase from a lactic acid bacterium, Leuconostoc mesenteroides subsp. sake NBRC 102480. Amino Acids 51, 331–343 (2019). https://doi.org/10.1007/s00726-018-2671-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-018-2671-y