Abstract
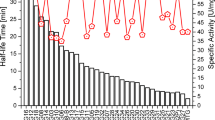
The thermostability of microbial transglutaminase (MTG) of Streptomyces mobaraensis was further improved by saturation mutagenesis and DNA-shuffling. High-throughput screening was used to identify clones with increased thermostability at 55°C. Saturation mutagenesis was performed at seven “hot spots”, previously evolved by random mutagenesis. Mutations at four positions (2, 23, 269, and 294) led to higher thermostability. The variants with single amino acid exchanges comprising the highest thermostabilities were combined by DNA-shuffling. A library of 1,500 clones was screened and variants showing the highest ratio of activities after incubation for 30 min at 55°C relative to a control at 37°C were selected. 116 mutants of this library showed an increased thermostability and 2 clones per deep well plate were sequenced (35 clones). 13 clones showed only the desired sites without additional point mutations and eight variants were purified and characterized. The most thermostable mutant (triple mutant S23V-Y24N-K294L) exhibited a 12-fold higher half-life at 60°C and a 10-fold higher half-life at 50°C compared to the unmodified recombinant wild-type enzyme. From the characterization of different triple mutants differing only in one amino acid residue, it can be concluded that position 294 is especially important for thermostabilization. The simultaneous exchange of amino acids at sites 23, 24, 269 and 289 resulted in a MTG-variant with nearly twofold higher specific activity and a temperature optimum of 55°C. A triple mutant with amino acid substitutions at sites 2, 289 and 294 exhibits a temperature optimum of 60°C, which is 10°C higher than that of the wild-type enzyme.




Similar content being viewed by others
Abbreviations
- FRAP-MTG-His6 :
-
Recombinant wild-type MTG
- MTG:
-
Microbial transglutaminase
- TG:
-
Transglutaminase
- t 1/2 :
-
Half-life
References
Ando H, Adachi M, Umeda K, Matsuura A, Nonaka M, Uchio R, Tanaka H, Motoki M (1989) Purification and characteristics of a novel transglutaminase derived from microorganisms. Agric Biol Chem 53(10):2613–2617
Beninati S, Piacentini M (2004) The transglutaminase family: an overview: minireview article. Amino Acids 26(4):367–372
Cadwell RC, Joyce GF (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl 2(1):28–33
Cadwell RC, Joyce GF (1994) Mutagenic PCR. PCR Methods Appl 3(6):S136–S140
Chambi H, Grosso C (2006) Edible films produced with gelatin and casein cross-linked with transglutaminase. Food Res Int 39:458–466
de Jong GA, Koppelman SJ (2002) Transglutaminase catalyzed reactions: impact on food applications. J Food Sci 67(8):2798–2806
Folk JE, Cole PW (1966) Transglutaminase: mechanistic features of the active site as determined by kinetic and inhibitor studies. Biochim Biophys Acta 122(2):244–264
Giver L, Gershenson A, Freskgard P-O, Arnold FH (1998) Directed evolution of a thermostable esterase. Proc Natl Acad Sci USA 95(22):12809–12813
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18(15):2714–2723
Kashiwagi T, Yokoyama K, Ishikawa K, Ono K, Ejima D, Matsui H, Suzuki E (2002) Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense. J Biol Chem 277(46):44252–44260
Longo MA, Combes D (1999) Thermostability of modified enzymes: a detailed study. J Chem Technol Biotechnol 74(1):25–32
Lorimer IAJ, Pastan I (1995) Random recombination of antibody single chain Fv sequences after fragmentation with DNasel in the presence of Mn2+. Nucleic Acids Res 23(15):3067–3068
Lowman HB, Wells JA (1993) Affinity maturation of human growth hormone by monovalent phage display. J Mol Biol 234(3):564–578
Marx C, Hertel T, Pietzsch M (2007) Soluble expression of a pro-transglutaminase from Streptomyces mobaraensis in Escherichia coli. Enzyme Microb Technol 40:1543–1550
Marx CK, Hertel TC, Pietzsch M (2008a) Purification and activation of a recombinant histidine-tagged pro-transglutaminase after soluble expression in E. coli and characterization of the active enzyme. Enzyme Microb Technol 42:568–575
Marx CK, Hertel TC, Pietzsch M (2008b) Random mutagenesis of a recombinant microbial transglutaminase for the generation of thermostable and heat sensitive variants. J Biotechnol 136:156–162
Maullu C, Raimondo D, Caboi F, Giorgetti A, Sergi M, Valentini M, Tonon G, Tramontano A (2009) Site-directed enzymatic PEGylation of the human granulocyte colony-stimulating factor. FEBS J 276(22):6741–6750
McLachlan MJ, Johannes TW, Zhao H (2008) Further improvement of phosphite dehydrogenase thermostability by saturation mutagenesis. Biotechnol Bioeng 99(2):268–274
Miyazaki K, Arnold FH (1999) Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J Mol Evol 49(6):716–720
O’Fagain C (2003) Enzyme stabilization—recent experimental progress. Enzyme Microb Technol 33(2–3):137–149
Oh J-H, Wang B, Field PD, Aglan HA (2004) Characteristics of edible films made from dairy proteins and zein hydrolysate cross-linked with transglutaminase. Int J Food Sci Technol 39(3):287–294
Pace CN, Schmid FX (1997) How to determine the molar absorption coefficient of a protein. Protein structure: a practical approach. IRL Press, Oxford
Park Y-M, Phi QT, Song B-H, Ghim S-Y (2009) Thermostability of chimeric cytidine deaminase variants produced by DNA shuffling. J Microbiol Biotechnol 19(12):1536–1541
Patzsch K, Riedel K, Pietzsch M (2010) Parameter optimization for protein film production using microbial transglutaminase. Biomacromolecules 11:896–903
Sato H (2002) Enzymatic procedure for site-specific pegylation of proteins. Adv Drug Deliv Rev 54(4):487–504
Schrodinger LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1
Shimba N, Shinohara M, Yokoyama K, Kashiwagi T, Ishikawa K, Ejima D, Suzuki E (2002) Enhancement of transglutaminase activity by NMR identification of its flexible residues affecting the active site. FEBS Lett 517(1–3):175–179
Sommer C, Hertel TC, Schmelzer CEH, Pietzsch M (2011a) Investigations on the activation of recombinant microbial pro-transglutaminase: in contrast to Proteinase K, Dispase removes the Histidine-tag. Amino Acids. doi:10.1007/s00726-011-1016-x (this issue)
Sommer C, Volk N, Pietzsch M (2011b) Model based optimization of the fed-batch production of a highly active transglutaminase variant in E. coli. Protein Expr Purif 77:9–19
Stemmer WPC (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature (London) 370(6488):389–391
Stratagene (2007) QuikChange site-directed Mutagenesis Kit. Manufacturer’s Manual
Yokoyama K, Utsumi H, Nakamura T, Ogaya D, Shimba N, Suzuki E, Taguchi S (2010) Screening for improved activity of a transglutaminase from Streptomyces mobaraensis created by a novel rational mutagenesis and random mutagenesis. Appl Microbiol Biotechnol 87(6):2087–2096
Yuan L, Kurek I, English J, Keenan R (2005) Laboratory-directed protein evolution. Microbiol Mol Biol Rev 69(3):373–392
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32(14):e115/111–e115/115
Acknowledgments
This research was supported by the “Fachagentur für Nachwachsende Rohstoffe” (FNR, BMELV, Germany, Project Code 22021807).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Buettner, K., Hertel, T.C. & Pietzsch, M. Increased thermostability of microbial transglutaminase by combination of several hot spots evolved by random and saturation mutagenesis. Amino Acids 42, 987–996 (2012). https://doi.org/10.1007/s00726-011-1015-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-1015-y