Abstract
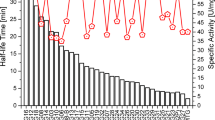
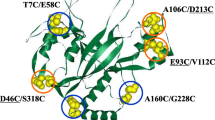
Microbial transglutaminase (MTG) has been used extensively in academic research and the food industries through its cross-linking or posttranslational modification of proteins. Two enzyme engineering approaches were applied to improve MTG activity. One is a novel method of rational mutagenesis, called water-accessible surface hot-space region-oriented mutagenesis (WASH-ROM). One hundred and fifty-one point mutations were selected at 40 residues, bearing high solvent-accessibility surface area, within a 15 Å space from the active site Cys64. Among them, 32 mutants showed higher specific activity than the wild type. The other is a random mutagenesis of the whole region of the MTG gene, coupled with a new plate assay screening system, using Corynebacterium Expression System CORYNEX®. This in vivo system allowed us to readily distinguish the change in enzymatic activity by monitoring the intensity of enzymatic reaction-derived color zones surrounding recombinant cells. From the library of 24,000 mutants, ten were finally selected as beneficial mutants exhibiting higher specific activity than the wild type. Furthermore, we found that Ser199Ala mutant with additional N-terminal tetrapeptide showed the highest specific activity (1.7 times higher than the wild type). These various beneficial positions leading to increased specific activity of MTG were identified to achieve further enzyme improvements.




Similar content being viewed by others
References
Ando H, Adachi M, Umeda K, Matsuura A, Nonaka M, Uchio R, Tanaka H, Motoki M (1989) Purification and characterization of a novel transglutaminase derived from microorganisms. Agric Biol Chem 53:2613–2617
Beninati S, Bergamini CM, Piacentini M (2009) An overview of the first 50 years of transglutaminase research. Amino Acids 36:591–598
Date M, Yokoyama K, Umezawa Y, Matsui H, Kikuchi Y (2003) Production of native-type Streptoverticillium mobaraense transglutaminase in Corynebacterium glutamicum. Appl Environ Microbiol 69:3011–3014
Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M (1995) The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J Comp Chem 16:273–284
Fontana A, Spolaore B, Mero A, Veronese FM (2008) Site-specific modification and PEGylation of pharmaceutical proteins mediated by transglutaminase. Adv Drug Deliv Rev 60:13–28
Ichinose A, Hendrickson LE, Fujikawa K, Davie EW (1986) Amino acid sequence of the subunit of human factor XIII. Biochemistry 25:6900–6906
Ikura K, Nasu T, Yokota H, Tsuchiya Y, Sasaki R, Chiba H (1988) Amino acid sequence of guinea pig liver transglutaminase from its cDNA sequence. Biochemistry 27:2898–2905
Kawajiri H, Ide H, Motoki M, Shimonishi Y (1993) Primary structure of microbial transglutaminase from Streptoverticillium sp. strain s-8112. J Biol Chem 268:11565–11572
Kashiwagi T, Yokoyama K, Ishikawa K, Ono K, Ejima D, Matui H, Suzuki E (2002) Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense. J Biol Chem 277:44252–44260
Kikuchi Y, Date M, Yokoyama K, Umezawa Y, Matsui H (2003) Secretion of active-form Streptoverticillium mobaraense transglutaminase by Corynebacterium glutamicum: processing of the pro-transglutaminase by a cosecreted subtilisin-like protease from Streptomyces albogriseolus. Appl Environ Microbiol 69:358–366
Kuraishi C, Sakamoto J, Soeda T (1996) The usefulness of transglutaminase for food processing. In: Takeoka GR, Teranishi R, Williams PJ, Kobayashi A (eds) Biotechnology for improved foods and flavors, ACS Symposium Series 637. American Chemical Society, Washington, DC, pp 29–38
Liebl W, Bayerl A, Schein B, Stillner U, Schleifer KH (1989) High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett 53:299–303
Marx CK, Hertel TC, Pietzsch M (2008) Random mutagenesis of a recombinant microbial transglutaminase for the generation of thermostable and heat-sensitive variants. J Biotechnol 136:156–162
Motoki M, Seguro K (1998) Transglutaminase and its use for food processing. Trends Food Sci Technol 9:204–210
Nonaka M, Tanaka H, Okiyama A, Motoki M, Ando H, Umeda K, Matsuura A (1989) Polymerization of several proteins by calcium-independent transglutaminase derived from microorganisms. Agric Biol Chem 53:2619–2623
Pasternack R, Dorsch S, Otterbach JT, Robenek IR, Wolf S, Fuchsbauer HL (1998) Bacterial pro-transglutaminase from Streptoverticillium mobaraense: purification, characterization and sequence of zymogen. Eur J Biochem 257:570–576
Sakamoto H, Nonaka M, Motoki M (1993) Calcium-independent transglutaminase derived from a microorganism: its characteristic and capability in protein crosslinking and gel formation. Food Hydrocoll 8:383–386
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Tagami U, Shimba N, Nakamura M, Yokoyama K, Suzuki E, Hirokawa T (2009) Substrate specificity of microbial transglutaminase as revealed by three-dimensional docking simulation and mutagenesis. Protein Eng Des Sel 22:747–752
Taguchi S, Tsuge T (2008) Natural polyester-related proteins: structure, function, evolution and engineering. In Lutz S, Bornschuer UT (eds) Protein engineering handbook. Wiley, Weinheim, pp 877–914
Taguchi S, Ozaki A, Momose H (1998) Engineering of a cold-adapted protease by sequential random mutagenesis and a screening system. Appl Environ Microbiol 64:492–495
Thacher SM (1989) Purification of keratinocyte transglutaminase and its expression during squamous differentiation. J Invest Dermatol 92:578–584
Umezawa Y, Yokoyama K, Kikuchi Y, Date M, Ito K, Yoshimoto T, Matsui H (2004) Novel Prolyl Tri/Tetra-Peptidyl aminopeptidase from Streptomyces mobaraensis: substrate specificity and enzyme gene cloning. J Biochem 136:293–300
Washizu K, Ando K, Koikeda S, Hirose S, Matsuura A, Takagi H, Motoki M, Takeuchi K (1994) Molecular cloning of the gene for microbail transglutaminase from streptoverticillium and its expression in Streptomyces lividans. Biosci Biotech Biochem 58:82–87
Yasueda H, Nakanishi K, Kumazawa Y, Nagase K, Motoki M, Matui H (1995) Tissue-type transglutaminase from red sea bream (Pagrus major) sequence analysis of the cDNA and functional expression in Escherichia coli. Eur J Biochem 232:411–419
Yokoyama K, Nakamura N, Saguaro K, Kubota K (2000) Overproduction of microbial transglutaminase in Escherichia coli, in vitro refolding, and characterization of the refolded form. Biosci Biotechnol Biochem 64:1263–1270
Yokoyama K, Ono K, Ohtsuka T, Nakamura N, Seguro K, Ejima D (2002) In vitro refolding process of urea-denatured microbial transglutaminase without pro-peptide sequence. Protein Expr Purif 26:329–335
Yokoyama K, Nio N, Kikuchi Y (2004) Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol 64:447–454
Zhu Y, Tramper J (2008) Novel applications for microbial transglutaminase beyond food processing. Trends Biotechnol 26:559–565
Zotzel J, Keller P, Fuchsbauer HL (2003a) Transglutaminase from Streptomyces mobaraensis is activated by an endogenous metalloprotease. Eur J Biochem 270:3214–3222
Zotzel J, Pasternack R, Pelzer C, Ziegert D, Mainusch M, Fuchsbauer HL (2003b) Activated transglutaminase from Streptomyces mobaraensis is processed by a tripeptidyl aminopeptidase in the final step. Eur J Biochem 270:4149–4155
Acknowledgments
We thank Dr. Kashiwagi and Dr. Nio for helpful discussions. We also thank Dr. Onoe and Ms. Tagami for technical assistance. This work was done by in part of the finance from the Global Center of Excellence Program (Project No. B01: Catalysis as the Basis for Innovation in Materials Science) from the Ministry of Education, Culture, Sports, Science, and Technology-Japan. Pacific Edit reviewed the manuscript prior to submission.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yokoyama, K., Utsumi, H., Nakamura, T. et al. Screening for improved activity of a transglutaminase from Streptomyces mobaraensis created by a novel rational mutagenesis and random mutagenesis. Appl Microbiol Biotechnol 87, 2087–2096 (2010). https://doi.org/10.1007/s00253-010-2656-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2656-6