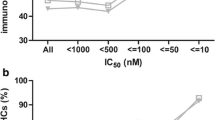
Abstract
Since CD8+ T cell response is crucial to combat intracellular infections and cancer, identification of class I HLA binding peptides is of immense clinical value. The experimental identification of such peptides is protracted and laborious. Exploiting in silico tools to discover such peptides is an attractive alternative. However, this approach needs a thorough assessment before its elaborate application. We have adopted a reverse approach to evaluate the reliability of eight different servers (inclusive of 55 predictors) by exploiting experimentally proven data. A comprehensive data set of more than 960 peptides was employed to test the efficacy of the programs. We have validated commonly used strategies to predict peptides that bind to seven most prevalent HLA class I alleles. We conclude that four of the eight servers are more adept in predictions. Although the overall predictions for class I MHC binders were superior to class II MHC binders, individual predictors for different alleles belonging to the same program were highly variable in their efficiencies. We have also addressed whether a consensus approach can yield better prediction efficiency. We observed that combining the results from different in silico programs could not increase the efficiency significantly.







Similar content being viewed by others
Abbreviations
- HLA:
-
Human leukocyte antigen
- MHC:
-
Major histocompatibility complex
- SVM:
-
Support vector machine
- ANN:
-
Artificial neural networks
References
Baldi P, Brunak S, Chauvin Y, Andersen CA et al (2000) Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 16:412–424
Bhasin M, Raghava GP (2007) A hybrid approach for predicting promiscuous MHC class I restricted T cell epitopes. J Biosci 32:31–42
Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A (2005) Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics 57(5):304–314
Burrows SR, Rossjohn J, McCluskey J (2006) Have we cut ourselves too short in mapping CTL epitopes? Trends Immunol 27:11–16
Donnes P, Kohlbacher O (2006) SVMHC: a server for prediction of MHC-binding peptides. Nucleic Acids Res 34:W194–W197
Garcia KC, Teyton L, Wilson IA (1999) Structural basis of T cell recognition. Annu Rev Immunol 17:369–397
Gowthaman U, Agrewala JN (2008) In silico tools for predicting peptides binding to HLA-class II molecules: more confusion than conclusion. J Prot Res 7:154–163
Gowthaman U, Agrewala JN (2009) In silico methods for predicting T-cell epitopes: Dr Jekyll or Mr Hyde? Expert Rev Proteomics 6:527–537
Harrison LC et al (1997) A peptide-binding motif for I-A(g7), the class II major histocompatibility complex (MHC) molecule of NOD and Biozzi AB/H mice. J Exp Med 185:1013–1021
Hoof I, Peters B, Sidney J, Pedersen L, Sette A, Lund O, Buus S, Nielsen M (2009) NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13
Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V (2008) Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol 9:8
Lundegaard C, Lambert K, Harndah M, Buus S, Lund O, Nielsen M (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512
MacNamara A, Kadolsky U, Bangham CR, Asquith B (2009) T-cell epitope prediction: rescaling can mask biological variation between MHC molecules. PLoS Comput Biol 5:e1000327
Parmiani G, De Filippo A, Novellino L, Castelli C (2007) Unique human tumor antigens: immunobiology and use in clinical trials. J Immunol 178:1975–1979
Peters B et al (2005) The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol 3:e91
Purcell AW, McCluskey J, Rossjohn J (2007) More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov 6:404–414
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50(3–4):213–219
Reche PA, Reinherz EL (2003) Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331(3):623–641
Reche PA, Glutting JP, Reinherz EL (2002) Prediction of MHC class I binding peptides using profile motifs. Human Immunol 63:701–709
Reche PA, Glutting JP, Zhang H, Reinherz EL (2004) Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56:405–419
Robinson J et al (2009) The IMGT/HLA database. Nucleic Acids Res 37:D1013–D1017
Stern LJ, Wiley DC (1994) Antigen peptide binding by class I and class II histocompatibility proteins. Structure 2:245–251
Tong JC, Tan TW, Ranganathan S (2006) Methods and protocols for prediction of immunogenic epitopes. Brief Bioinform 8:96–108
Topalian SL et al (1996) Melanoma-specific CD4+ T cells recognize nonmutated HLA-DR-restricted tyrosinase epitopes. J Exp Med 183:1965–1971
Toseland CP, Clayton DJ, McSparron H, Hemsley SL, Blythe MJ, Paine K, Doytchinova IA, Guan P, Hattotuwagama CK, Flower DR (2005) AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data. Immunome Res 1(1):4
Toseland CP, Clayton DJ, McSparron H, Hemsley SL, Blythe MJ, Paine K, Trost B, Bickis M, Kusalik A (2007) Strength in numbers: achieving greater accuracy in MHC-I binding prediction by combining the results from multiple prediction tools. Immunome Res 3:5
Wang P et al (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4:e1000048
Zhang Q et al (2008) Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res 36:W513–W518
Zinkernagel RM, Doherty PC (1974) Restriction of in vitro T-cell mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semi allogeneic system. Nature 248:701–702
Acknowledgments
The authors are thankful to Dr. Balvinder Singh, Institute of Microbial Technology, Chandigarh for valuable suggestions and discussions and Priya Gowthaman for her help in manuscript preparation. UG is a recipient of Junior Research Fellowship of Department of Biotechnology. CSB is a recipient of Junior Research Fellowship of Council of Scientific and Industrial Research. The authors also wish to thank Department of Biotechnology and Council of Scientific and Industrial Research, India for financial support.
Conflict of interest statement
The authors declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
U. Gowthaman and S. B. Chodisetti have contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
726_2010_579_MOESM1_ESM.pdf
Variability in the prediction efficiency for different alleles in a single program. Matthew’s correlation coefficients for different alleles from a single program is depicted as bar diagrams for Rankpep (A), ComPred (B), NetMHC (C), NetMHCPan (D), ARB-Matrix (E), IEDB-ANN (F), SVMHC-SYFPEITHI (G), SVMHC-MHCPep (H) (PDF 33 kb)
Rights and permissions
About this article
Cite this article
Gowthaman, U., Chodisetti, S.B., Parihar, P. et al. Evaluation of different generic in silico methods for predicting HLA class I binding peptide vaccine candidates using a reverse approach. Amino Acids 39, 1333–1342 (2010). https://doi.org/10.1007/s00726-010-0579-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-010-0579-2