Abstract
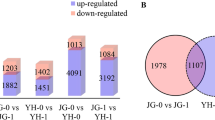
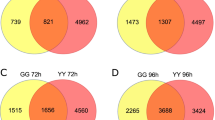
Rice sheath blight (ShB) disease, caused by the fungal pathogen Rhizoctonia solani AG1-IA, is one of the devastating diseases and causes severe yield losses all over the world. No completely resistant germplasm is known till now, and as a result, the progress in resistance breeding is unsatisfactory. Basic studies to identify candidate genes, QTLs, and to better understand the host–pathogen interaction are also scanty. In this study, we report the identification of a new ShB-tolerant rice germplasm, CR 1014. Further, we investigated the basis of tolerance by exploring the disease responsive differentially expressed transcriptome and comparing them with that of a susceptible variety, Swarna-Sub1. A total of 815 and 551 genes were found to be differentially regulated in CR 1014 and Swarna-Sub1, respectively, at two different time points. The result shows that the ability to upregulate genes for glycosyl hydrolase, secondary metabolite biosynthesis, cytoskeleton and membrane integrity, the glycolytic pathway, and maintaining photosynthesis make CR 1014 a superior performer in resisting the ShB pathogen. We discuss several putative candidate genes for ShB resistance. The present study, for the first time, revealed the basis of ShB tolerance in the germplasm CR1014 and should prove to be particularly valuable in understanding molecular response to ShB infection. The knowledge could be utilized to devise strategies to manage the disease better.





Similar content being viewed by others
References
Bal A, Samal P, Chakraborti M, Mukherjee AK, Ray S, Molla KA, Behera L, Samal R, Sarangi S, Sahoo P, Behera M, Lenka S, Azharudheen TPM, Khandual A, Kar MK (2020) Stable quantitative trait locus (QTL) for sheath blight resistance from rice cultivar CR 1014. Euphytica 216. https://doi.org/10.1007/s10681-020-02702-x
Bera S, Purkayastha RP (1999) Multicomponent coordinated defence response of rice to Rhizoctonia solani causing sheath blight. Curr Sci 76(10):1376–1384
Bhaktavatsalam G, Satyanarayana K, Reddy APK, John VT (1978) Evaluation for sheath blight resistance in rice. Int Rice Res Newsl Int Rice Res Inst 3:9–10
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: Architecture and applications. BMC Bioinformatics 10:1–9. https://doi.org/10.1186/1471-2105-10-421
Channamallikarjuna V, Sonah H, Prasad M, Rao GJN, Chand S, Upreti HC, Singh NK, Sharma TR (2010) Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol Breed 25:155–166. https://doi.org/10.1007/s11032-009-9316-5
Chen G, Zou Y, Hu J, Ding Y (2018a) Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19:1–14. https://doi.org/10.1186/s12864-018-5088-9
Chen X, Chen H, Yuan JS, Köllner TG, Chen Y, Guo Y, Zhuang X, Chen X, Zhang YJ, Fu J, Nebenführ A, Guo Z, Chen F (2018b) The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol J 16:1778–1787. https://doi.org/10.1111/pbi.12914
Conesa A, Götz S, García-Gómez JM et al (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Cunha SR, Mohler PJ (2009) Ankyrin protein networks in membrane formation and stabilization. J Cell Mol Med 13:4364–4376. https://doi.org/10.1111/j.1582-4934.2009.00943.x
Danson J, Wasano K, Nose A (2000) Infection of rice plants with the sheath blight fungus causes an activation of pentose phosphate and glycolytic pathways. Eur J Plant Pathol 106:555–561
Dao TTH, Linthorst HJM, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412. https://doi.org/10.1007/s11101-011-9211-7
Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK (1999) Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet 98:1138–1145. https://doi.org/10.1007/s001220051178
Dhakarey R, Kodackattumannil Peethambaran P, Riemann M (2016) Functional analysis of jasmonates in rice through mutant approaches. Plants 5:267–281. https://doi.org/10.3390/plants5010015
El-Zahaby HM, Gullner G, Kiraly Z (1995) Effects of powdery mildew infection of barley on the ascorbate-glutathione cycle and other antioxidants in different host-pathogen interactions. Phytopathology 85:1225–1230
Ghosh S, Kanwar P, Jha G (2017) Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani. Sci Rep 7:41610. https://doi.org/10.1038/srep41610
Gullner G, Komives T, Király L, Schröder P (2018) Glutathione S-transferase enzymes in plant-pathogen interactions. Front Plant Sci 871:1–19. https://doi.org/10.3389/fpls.2018.01836
Guo WJ, Ho THD (2008) An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147:1710–1722. https://doi.org/10.1104/pp.108.120238
Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW (2012) Plant disease resistance genes: current status and future directions. Physiol Mol Plant Pathol 78:51–65
Han Y, Ding T, Su B, Jiang H (2016) Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int J Mol Sci 17. https://doi.org/10.3390/ijms17020161
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444:11–25. https://doi.org/10.1042/BJ20120245
Henty-Ridilla JL, Shimono M, Li J, Chang JH, Day B, Staiger CJ (2013) The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog 9. https://doi.org/10.1371/journal.ppat.1003290
Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA 100:4921–4926. https://doi.org/10.1073/pnas.0736939100
IRRI (2002) Standard evaluation system for rice. International Rice Research Institute, Los Banos, Manila
Jia Y, Correa-Victoria F, McClung A, Zhu L, Liu G, Wamishe Y, Xie J, Marchetti MA, Pinson SRM, Rutger JN, Correll JC (2007) Rapid determination of rice cultivar responses to the sheath blight pathogen Rhizoctonia solani using a micro-chamber screening method. Plant Dis 91:485–489. https://doi.org/10.1094/PDIS-91-5-0485
Kalpana K, Maruthasalam S, Rajesh T, Poovannan K, Kumar KK, Kokiladevi E, Raja JAJ, Sudhakar D, Velazhahan R, Samiyappan R, Balasubramanian P (2006) Engineering sheath blight resistance in elite indica rice cultivars using genes encoding defense proteins. Plant Sci 170:203–215. https://doi.org/10.1016/j.plantsci.2005.08.002
Karmakar S, Molla KA, Chanda PK, Sarkar SN, Datta SK, Datta K (2016) Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 243:115–130
Karmakar S, Datta K, Molla KA, Gayen D, Das K, Sarkar SN, Datta SK (2019) Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and accumulation of novel proteins potentially involved in defence against Rhizoctonia solani. Sci Rep 9:1–16. https://doi.org/10.1038/s41598-019-46885-3
Karunanithi PS, Zerbe P (2019) Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front Plant Sci 10:1–23. https://doi.org/10.3389/fpls.2019.01166
Kim Y, Hao J, Gautam Y, Mersha TB, Kang M (2018) DiffGRN: Differential gene regulatory network analysis. Int J Data Min Bioinform 20:362–379. https://doi.org/10.1504/IJDMB.2018.094891
Koga J, Shimura M, Oshima K, Ogawa N, Yamauchi T, Ogasawara N (1995) Phytocassanes A, B, C and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 51:7907–7918
Lee HA, Yeom SI (2015) Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief Funct Genomics 14:233–242. https://doi.org/10.1093/bfgp/elv012
Li Z, Pinson SRM, Marchetti MA, Stansel JW, Park WD (1995) Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor Appl Genet 91:382–388. https://doi.org/10.1007/BF00220903
Maeda S, Dubouzet JG, Kondou Y, Jikumaru Y, Seo S, Oda K, Matsui M, Hirochika H, Mori M (2019) The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Sci Rep 9:1–14. https://doi.org/10.1038/s41598-018-37365-1
Mafu S, Jia M, Zi J, Morrone D, Wu Y, Xu M, Hillwig ML, Peters RJ (2016) Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proc Natl Acad Sci USA 113:2526–2531. https://doi.org/10.1073/pnas.1512096113
Molla KA, Karmakar S, Chanda PK, Ghosh S, Sarkar SN, Datta SK, Datta K (2013) Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol Plant Pathol 14:910–922. https://doi.org/10.1111/mpp.12055
Molla KA, Karmakar S, Chanda PK, Sarkar SN, Datta SK, Datta K (2016) Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci 250:105–114
Molla KA, Azharudheen TPM, Ray S, Sarkar S, Swain A, Chakraborti M, Vijayan J, Singh ON, Baig MJ, Mukherjee AK (2019) Novel biotic stress responsive candidate gene based SSR (cgSSR) markers from rice. Euphytica 215:17
Molla KA, Karmakar S, Molla J, Bajaj P, Varshney RK, Datta SK, Datta K (2020) Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol J 18:895–915. https://doi.org/10.1111/pbi.13312
Mukherjee AK, Carp MJ, Zuchman R, Ziv T, Horwitz BA, Gepstein S (2010) Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J Proteome 73:709–720. https://doi.org/10.1016/j.jprot.2009.10.005
Musembi Mutuku J, Nose A (2012) Changes in the contents of metabolites and enzyme activities in rice plants responding to Rhizoctonia solani Kuhn infection: activation of glycolysis and connection to phenylpropanoid pathway. Plant Cell Physiol 53:1017–1032. https://doi.org/10.1093/pcp/pcs047
Mutuku M, Nose A (2010) Rhizoctonia solani infection in two rice lines increases mRNA expression of metabolic enzyme genes in glycolytic, oxidative pentose phosphate pathways and secondary metabolism. Trop Agric Dev 54:119–131
Nelson B, Helms T, Christianson T, Kural I (1996) Characterization and pathogenicity of Rhizoctonia from soybean. Plant Dis 80:74
Okubara PA, Dickman MB, Blechl AE (2014) Molecular and genetic aspects of controlling the soilborne necrotrophic pathogens Rhizoctonia and Pythium. Plant Sci 228:61–70. https://doi.org/10.1016/j.plantsci.2014.02.001
Pan X, Zou J, Chen Z, Lu J, Yu H, Li H, Wang Z, Pan X, Rush MC, Zhu L (1999) Tagging major quantitative trait loci for sheath blight resistance in a rice variety, Jasmine 85. Chin Sci Bull 44:1783–1789. https://doi.org/10.1007/BF02886159
Park DS, Sayler RJ, Hong YG, Nam MH, Yang Y (2008) A method for inoculation and evaluation of rice sheath blight disease. Plant Dis 92:25–29. https://doi.org/10.1094/PDIS-92-1-0025
Park HL, Yoo Y, Hahn TR, Bhoo S, Lee SW, Cho MH (2014) Antimicrobial activity of uv-induced phenylamides from rice leaves. Molecules 19:18139–18151. https://doi.org/10.3390/molecules191118139
Peng X, Wang H, Jang JC, Xiao T, He H, Jiang D, Tang X (2016) OsWRKY80-OsWRKY4 Module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice 9:63. https://doi.org/10.1186/s12284-016-0137-y
Prakasham V, Ladhalakshmi D, Laha GS, Krishnaveni D, Sheshu Madhav M, Jyothi Badri, Srinivas Prasad, M, Viraktamath BC (2013) Sheath blight of rice and its management. Technical Bulletin No. 72, Directorate of Rice Research (ICAR), Rajendranagar, Hyderabad-500 030,A.P., India, 58 pp.
Reape TJ, Molony EM, McCabe PF (2008) Programmed cell death in plants: distinguishing between different modes. J Exp Bot 59:435–444. https://doi.org/10.1093/jxb/erm258
Sato H, Ideta O, Ando I, Kunihiro Y, Hirabayashi H, Iwano M, Miyasaka A, Nemoto H, Imbe T (2004) Mapping QTLs for sheath blight resistance in the rice line WSS2. Breed Sci 54:265–271
Sayari M, Babaeizad V, Ghanbari MAT, Rahimian H (2014) Expression of the pathogenesis related proteins, nh-1, pal, and lipoxygenase in the Iranian Tarom and khazar rice cultivars, in reaction to Rhizoctonia solani - the causal agent of rice sheath blight. J Plant Prot Res 54:36–43. https://doi.org/10.2478/jppr-2014-0006
Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79:659–678. https://doi.org/10.1111/tpj.12436
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Sharma A, McClung AM, Pinson SRM et al (2009) Genetic mapping of sheath blight resistance QTLs within tropical Japonica rice cultivars. Crop Sci 49:256–264. https://doi.org/10.2135/cropsci2008.03.0124
Shi W, Zhao SL, Liu K, Sun YB, Ni ZB, Zhang GY, Tang HS, Zhu JW, Wan BJ, Sun HQ, Dai JY (2020) Comparison of leaf transcriptome in response to Rhizoctonia solani infection between resistant and susceptible rice cultivars. BMC Genomics 21(1):1–6
Singh DK, McNellis TW (2011) Fibrillin protein function: the tip of the iceberg? Trends Plant Sci 16:432–441
Srinivasachary WL, Savary S (2011) Resistance to rice sheath blight (Rhizoctonia solani Kühn) [teleomorph: Thanatephorus cucumeris (A.B. Frank) Donk.] disease: current status and perspectives. Euphytica 178:1–22. https://doi.org/10.1007/s10681-010-0296-7
Sruthilaxmi CB, Babu S (2020) Proteome responses to individual pathogens and abiotic conditions in rice seedlings. Phytopathology 110(7):1326–1341. https://doi.org/10.1094/PHYTO-11-19-0425-R
Steinmüller D, Tevini M (1985) Composition and function of plastoglobuli. Planta 163:201–207
Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21:3315–3325
Taheri P, Tarighi S (2010) Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J Plant Physiol 167:201–208
Tevini MSD (1985) Composition and function of plastoglobuli. Planta 163:91–96
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. https://doi.org/10.1111/j.1365-313X.2004.02016.x
Vidhyasekaran P, Ruby Ponmalar T, Samiyappan R et al (1997) Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen. Phytopathology 87:1258–1263. https://doi.org/10.1094/PHYTO.1997.87.12.1258
Wang R, Lu L, Pan X, Hu Z, Ling F, Yan Y, Liu Y, Lin Y (2015) Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol Biol 87:181–191
Xu Y, Liu F, Zhu S, Li X (2018) The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and arabidopsis. Front Plant Sci 9:1–13. https://doi.org/10.3389/fpls.2018.01033
Xue X, Cao ZX, Zhang XT, Wang Y, Zhang YF, Chen ZX, Pan XB, Zuo SM (2016) Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis 100:1634–1642. https://doi.org/10.1094/PDIS-11-15-1372-RE
Yadav S, Anuradha G, Kumar RR et al (2015) Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). Springerplus 4. https://doi.org/10.1186/s40064-015-0954-2
Yuan Z, Zhang Y, Xu G, Bi D, Qu H, Zou X, Gao X, Yang H, He H, Wang X, Bao J, Zuo S, Pan X, Zhou B, Wang GL, Qu S (2018) Comparative transcriptome analysis of Rhizoctonia solani-resistant and susceptible rice cultivars reveals the importance of pathogen recognition and active immune responses in host resistance. J Plant Biol 61:143–158. https://doi.org/10.1007/s12374-017-0209-6
Zaynab M, Fatima M, Abbas S, Sharif Y, Umair M, Zafar MH, Bahadar K (2018) Role of secondary metabolites in plant defense against pathogens. Microb Pathog 124:198–202
Zhang Y, Xia R, Kuang H, Meyers BC (2016) The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol 33:2692–2705. https://doi.org/10.1093/molbev/msw154
Zhang J, Chen L, Fu C, Wang L, Liu H, Cheng Y, Li S, Deng Q, Wang S, Zhu J, Liang Y, Li P, Zheng A (2017) Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front Plant Sci 8:1–18. https://doi.org/10.3389/fpls.2017.01422
Zhou N, Tootle TL, Glazebrook J (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11:2419–2428. https://doi.org/10.1105/tpc.11.12.2419
Zou JH, Pan XB, Chen ZX, Xu JY, Lu JF, Zhai WX, Zhu LH (2000) Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor Appl Genet 101:569–573. https://doi.org/10.1007/s001220051517
Author information
Authors and Affiliations
Contributions
PS, AB, HS, AK, PRS, and MLB carried out net house screening; AKM and MKK conceptualized and designed the experiments. AKM supervised the work. PS performed the experiments. PS, KM, and SR analyzed the data; PS and KM wrote the manuscript and prepared the figures. SR, AI, SJ, and LB carried out the bioinformatics and statistical analyses. MC and MKK critically reviewed the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Handling Editor: Néstor Carrillo
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
RNA-Seq analysis of two contrasting rice varieties reveals that the genes involved in cytoskeleton and membrane integrity, photosynthesis, and secondary metabolites play a crucial role in sheath blight disease tolerance.
Rights and permissions
About this article
Cite this article
Samal, P., Molla, K.A., Bal, A. et al. Comparative transcriptome profiling reveals the basis of differential sheath blight disease response in tolerant and susceptible rice genotypes. Protoplasma 259, 61–73 (2022). https://doi.org/10.1007/s00709-021-01637-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-021-01637-x