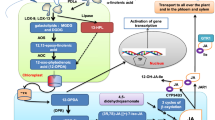
Abstract
Medicinal secondary metabolites (salvianolic acids and tanshinones) are valuable natural bioactive compounds in Salvia miltiorrhiza and have widespread applications. Improvement of medicinal secondary metabolite accumulation through biotechnology is necessary and urgent to satisfy their increasing demand. Herein, it was demonstrated that the overexpression of the transcription factor Arabidopsis thaliana-enhanced drought tolerance 1 (AtEDT1) could affect medicinal secondary metabolite accumulation. In this study, we observed that the transgenic lines significantly conferred drought tolerance phenotype. Meanwhile, we found that the overexpression of AtEDT1 promoted root elongation in S. miltiorrhiza. Interestingly, we also found that the overexpression of AtEDT1 determined the accumulation of salvianolic acids, such as rosmarinic acid, lithospermic acid, salvianolic acid B, and total salvianolic acids due to the induction of the expression levels of salvianolic acid biosynthetic genes. Conversely, S. miltiorrhiza plants overexpressing the AtEDT1 transgene showed a decrease in tanshinone synthesis. Our results demonstrated that the overexpression of AtEDT1 significantly increased the accumulation of salvianolic acids in S. miltiorrhiza. Further studies are required to better elucidate the functional role of AtEDT1 in the regulation of phytochemical compound synthesis.







Similar content being viewed by others
References
Anderegg WR, Anderegg LD (2013) Hydraulic and carbohydrate changes in experimental drought-induced mortality of saplings in two conifer species. Tree Physiol 33:252–260
Anderegg WR, Berry JA, Field CB (2012) Linking definitions, mechanisms, and modeling of drought-induced tree death trends. Plant Sci 17:693–700
Cai XT, Xu P, Wang Y, Xiang CB (2015) Activated expression of AtEDT1/HDG11 promotes lateral root formation in Arabidopsis mutant edt1 by upregulating jasmonate biosynthesis. J Integr Plant Biol 57:1017–1030
Cao YJ, Wei Q, Liao Y, Song HL, Li X, Xiang CB, Kuai BK (2009) Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep 28:579–588
Cao J, Hu J, Wei J, Li B, Zhang M, Xiang C, Li P (2015) Optimization of micellar electrokinetic chromatography method for the simultaneous determination of seven hydrophilic and four lipophilic bioactive components in three Salvia species. Molecules 20:15304–15318
Cheng Q, He Y, Li G, Liu Y, Gao W, Huang L (2013) Effects of combined elicitors on tanshinone metabolic profiling and SmCPS expression in Salvia miltiorrhiza hairy root cultures. Molecules 18:7473–7485
Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, John HS, Song J, Chen S, Huang L (2015) Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant Physiol. pp. 00695.02015
Doyle JJT, Doyle JL (1990) Isolation of plant DNA fresh tissue., p 12
Duan H, Duursma RA, Huang G, Smith RA, Choat B, O’Grady AP, Tissue DT (2014) Elevated [CO2] does not ameliorate the negative effects of elevated temperature on drought-induced mortality in Eucalyptus radiata seedlings. Plant Cell Environ 37:1598–1613
Gao W, Sun HX, Xiao H, Cui G, Hillwig ML, Jackson A, Wang X, Shen Y, Zhao N, Zhang L (2014) Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics 15:1
Hatfield MJ, Tsurkan LG, Hyatt JL, Edwards CC, Lemoff A, Jeffries C, Yan B, Potter PM (2013) Modulation of esterified drug metabolism by tanshinones from Salvia miltiorrhiza (“Danshen”). J Nat Prod 76:36–44
He M, Dijkstra FA (2014) Drought effect on plant nitrogen and phosphorus: a meta-analysis. New Phytol 204:924–931
Kai G, Xu H, Zhou C, Liao P, Xiao J, Luo X, You L, Zhang L (2011) Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab Eng 13:319–327
Kai G, Hao X, Cui L, Ni X, Zekria D, Wu JY (2014) Metabolic engineering and biotechnological approaches for production of bioactive diterpene tanshinones in Salvia miltiorrhiza. Biotechnol Adv.
Liu L, Jia J, Zeng G, Zhao Y, Qi X, He C, Guo W, Fan D, Han G, Li Z (2013) Studies on immunoregulatory and anti-tumor activities of a polysaccharide from Salvia miltiorrhiza Bunge. Carbohydr Polym 92:479–483
Liu Y, Yang SX, Cheng Y, Liu DQ, Zhang Y, Deng KJ, Zheng XL (2015) Production of herbicide-resistant medicinal plant salvia miltiorrhiza transformed with the Bar gene. Appl Biochem Biotechnol 177:1456–1465
Ma XH, Ma Y, Tang JF, He YL, Liu YC, Ma XJ, Shen Y, Cui GH, Lin HX, Rong QX (2015) The biosynthetic pathways of tanshinones and phenolic acids in salvia miltiorrhiza. Molecules 20:16235–16254
Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42:D142–D147
Palta JA, Chen X, Milroy SP, Rebetzke GJ, Dreccer MF, Watt M (2011) Large root systems: are they useful in adapting wheat to dry environments? Funct Plant Biol 38:347–354
Puértolas J, Ballester C, Elphinstone ED, Dodd IC (2014) Two potato (Solanum tuberosum) varieties differ in drought tolerance due to differences in root growth at depth. Funct Plant Biol 41:1107–1118
Ruan L, Chen L, Chen Y, He J, Zhang W, Gao Z, Zhang Y (2012) Expression of Arabidopsis HOMEODOMAIN GLABROUS 11 enhances tolerance to drought stress in transgenic sweet potato plants. J Plant Biol 55:151–158
Sgaramella V, Ehrlich SD (1978) Use of the T4 polynucleotide ligase in the joining of flush-ended DNA segments generated by restriction endonucleases. Eur J Biochem/FEBS 86:531–537
Shi M, Luo X, Ju G, Li L, Huang S, Zhang T, Wang H, Kai G (2016) Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J Agr Food Chem 64:2523
Song MC, Kim EJ, Kim E, Rathwell K, Nam SJ, Yoon YJ (2014) Microbial biosynthesis of medicinally important plant secondary metabolites. Nat Prod Rep 31:1497–1509
Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJ, Bouget FY, Ratsch G, Larrondo LF, Ecker JR, Hughes TR (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158:1431–1443
Wi SJ, Ji NR, Park KY (2012) Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic Phytophthora parasitica in tobacco plants. Plant Physiol 159:251–265
Xiao Y, Zhang L, Gao S, Saechao S, Di P, Chen J, Chen W (2011) The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PLoS One 6:e29713
Xu P, Cai X-T, Wang Y, Xing L, Chen Q, Xiang C-B (2014) HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J Exp Bot 65:4285, eru202
Xu Z, Peters RJ, Weirather J, Luo H, Liao B, Zhang X, Zhu Y, Ji A, Zhang B, Hu S (2015) Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 82:951–961
Xu H, Song J, Luo H, Zhang Y, Li Q, Zhu Y, Xu J, Li Y, Song C, Wang B (2016a) Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol Plant 9:949–952
Xu Z, Luo H, Ji A, Zhang X, Song J, Chen S (2016b) Global identification of the full-length transcripts and alternative splicing related to phenolic acid biosynthetic genes in Salvia miltiorrhiza. Front Plant Sci 7:100
Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20:1134–1151
Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C (2013) Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol 162:1378–1391
Yuan H, Chen L, Paliyath G, Sullivan A, Murr DP (2005) Characterization of microsomal and mitochondrial phospholipase D activities and cloning of a phospholipase D alpha cDNA from strawberry fruits. Plant Physiol Biochem 43:535–547
Zeppel MJ, Harrison SP, Adams HD, Kelley DI, Li G, Tissue DT, Dawson TE, Fensham R, Medlyn BE, Palmer A (2015) Drought and resprouting plants. New Phytol 206:583–589
Zhang Y, Yan YP, Wang ZZ (2010) The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza. J Agric Food Chem 58:12168–12175
Zhang Y, Yan YP, Wu YC, Hua WP, Chen C, Ge Q, Wang ZZ (2014) Pathway engineering for phenolic acid accumulations in Salvia miltiorrhiza by combinational genetic manipulation. Metab Eng 21:71–80
Zhao S, Zhang J, Tan R, Yang L, Zheng X (2015) Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J Exp Bot 66:7211, erv418
Zhou GJ, Wang W, Xie XM, Qin MJ, Kuai BK, Zhou TS (2014) Post-harvest induced production of salvianolic acids and significant promotion of antioxidant properties in roots of Salvia miltiorrhiza (Danshen). Molecules 19:7207–7222
Zhou Y, Sun W, Chen J, Tan H, Xiao Y, Li Q, Ji Q, Gao S, Chen L, Chen S (2016) SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci Rep 6:22852
Acknowledgements
We express our gratitude to Dr. Chengbin Xiang from the University of Science and Technology of China for providing the vector pCB2006-EDT1. The research was supported by the Fundamental Research Funds for the Central Universities (ZYGX2014J081) and the National Natural Science Foundation of China (31271420 and 31371682).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Handling Editor: Néstor Carrillo
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Contents of some components in the salvianolic acids and tanshinones biosynthesis pathway in wild type and transgenic lines determined by HPLC. ①rosmarinic acid, ②lithospermic acid, ③salvianolic acid B, ④dihydrotanshinone I, ⑤cryptotanshinone, ⑥tanshinone IIA. (PPTX 10978 kb)
Fig. S2
Biosynthesis of salvianolic acids and tanshinones in S. miltiorrhiza. Solid arrows indicate one step biosynthetic reaction, and dashed arrows indicate multiple-step reactions. E4P, erythose 4-phosphate; PEP, phosphoenolpyruvate; 4-HPPA, 4-Hydroxyphenylpyruvic acid; 4-HPLA, 4-hydroxyphenyllactic acid; G3P, glyceraldehyde 3-phosphate; HMG-CoA, 3-Hydroxy-3-methylglutary l-CoA; MVA, mevalonate; CDP-ME2P, 2-phospho-4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol; ME-cPP, 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGPP, geranylgeranyl diphosphate. (PPTX 8905 kb)
Table S1
(DOCX 21 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Sun, G., Zhong, Z. et al. Overexpression of AtEDT1 promotes root elongation and affects medicinal secondary metabolite biosynthesis in roots of transgenic Salvia miltiorrhiza . Protoplasma 254, 1617–1625 (2017). https://doi.org/10.1007/s00709-016-1045-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-016-1045-0