Abstract
Hydroxyurea (HU), an inhibitor of ribonucleotide reductase, prevents cells from progressing through S phase by depletion of deoxyribonucleoside triphosphates. Concurrently, disruption of DNA replication leads to double-strand DNA breaks. In root meristems of Vicia faba, HU triggers cell cycle arrest (preferentially in G1/S phase) and changes an overall metabolism by global activation of transcription both in the nucleoplasmic and nucleolar regions. High level of transcription is accompanied by an increase in the content of RNA polymerase II large subunit (POLR2A). Changes in transcription activation and POLR2A content correlate with posttranslational modifications of histones that play a role in opening up chromatin for transcription. Increase in the level of H4 Lys5 acetylation indicates that global activation of transcription following HU treatment depends on histone modifications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cell cycle transitions throughout interphase and mitosis are regulated by sophisticated metabolic pathways comprising diverse proteins. To ensure DNA integrity and correct functioning of important cellular processes, such as replication or mitotic division, cells take advantage of their evolutionary developed mechanisms called cell cycle checkpoints. Genotoxic stress caused by a variety of potential stressors (e.g., ultraviolet light, ionizing radiation, chemicals that affect DNA integrity) combined with disruption of checkpoint control functions may have a destructive impact on cellular functioning (Shackelford et. al. 1999; Bartek and Lukas 2001; Rybaczek and Kowalewicz-Kulbat 2011). Adequate response to DNA damage is possible owing to the presence of specific sensor kinases that constitute upstream factors to their effectors. Multidimensional character of cell cycle checkpoints allows not only to block phase-to-phase transitions but also, if necessary, to activate diverse genes and to trigger DNA repair processes (Jackson 2002; Yang et al. 2004). Replication fork stalling or DNA lesions are detected by two sensor kinases (PIKK family members), ATM (ataxia telangiectasia-mutated) and ATR (ATM and Rad 3-related) and their downstream factors—Chk1 and Chk2 kinases. Consequently, the activity of the latter proteins drives inhibitory phosphorylation of Cdc25 phosphatases, which become unable to activate cyclin and Cdk complexes, ultimately resulting in cell cycle arrest (Abraham 2001; Ricaud et al. 2007; McNeely et al. 2010). In turn, repair factors involved in homologous recombination or non-homologous end-joining are recruited to DNA lesions, owing to gamma phosphorylation of H2AX histones by sensor kinases (Rogakou et al. 1998; Hanakahi et al. 2000; Bassing et al. 2002). Molecular components of cell cycle checkpoints are well known, yet mutual relationships between these factors and other cellular proteins are still not clear.
Hydroxyurea (HU) is a well-known inhibitor of ribonucleotide reductase (RNR) which, by transformation of ribonucleotides to deoxyribonucleoside triphosphates, plays a fundamental role in establishing balanced quantities of precursors required for DNA synthesis and DNA repair systems (Roa et al. 2009; Koç et al. 2004; Alvino et al. 2007). Inhibition of RNR activity prevents cells from progressing through S phase by enhanced production of replication intermediates, including long ssDNA regions and inappropriate processing of the reversed forks, which leads to dsDNA breaks (DSB; Sogo et al. 2002). Moreover, HU triggers changes in an overall cellular metabolism resulting in an enhanced expression of diverse genes, e.g., γ-globin (Watanapokasin et al. 2006), adult β-globin in HeLA cells (Zhang et al. 2001), or SMN2 (survival motor neuron) in lymphoblastoid cell lines (Grzeschik et al. 2005). There are only few reports considering the effect of HU on transcription activation, and the molecular bases of this phenomenon are poorly understood. The results presented in this work indicate that in root meristems of Vicia faba, HU not only leads to cell cycle arrest but also triggers changes in cellular metabolic state manifested by global transcription activation correlated with an increased content of RNA polymerase II and histone H4 Lys5 acetylation (H4K5Ac).
Material and methods
Material
Seeds of V. faba subsp. minor var. Nadwiślański were sown on wet filter paper in Petri dishes and germinated for 3 days at room temperature in darkness. For experiments, seedlings with roots ranging from 1.5 to 2.0 cm in length were selected and incubated in water or 2.5 mM HU for 24 h.
Chemical agents
Hydroxyurea, pararosaniline, ethylenediaminetetra acetic acid, HEPES, phenylsulfonyl fluoride, dithiothreitol, Protease Inhibitor Cocktail (P-9599), Coomassie blue, DABCO (1,4-diazabicyclo [2.2.2] octane), and 4′,6-diamidino-2-phenylindole (DAPI) were supplied by Sigma, Triton X-100 and pectinase from Aspergillus niger by Fluka, cellulose Onozuka R-10 from Trichoderma viride by SERVA, pectolyase Y-23 by ICN, and acetic acid by Chempur. Click-iT® RNA Alexa Fluor® 488 Imaging Kit for visualization of RNA transcripts, NuPAGE® Novex® 4–12 % Bis-Tris gel, NuPAGE® Novex® 3–8 % Tris-Acetate gel, polyvinylidene fluoride membrane (0.2-μm pore size), and Chromogenic Western Blot Immunodetection Kit were supplied by Invitrogen. P-PER Plant Protein Extraction Kit was obtained from Pierce (Rochford, USA). Other chemicals were obtained from POCH S.A.
Feulgen staining and cytophotometry
Apical fragments of roots (1.5 cm long) were fixed in Carnoy’s mixture (ethanol/glacial acetic acid; 3:1, v/v) for 1 h. Following fixation, roots were rinsed three times in 96 % ethanol, rehydrated (70–30 % ethanol, distilled water), hydrolyzed in 4 M HCl (1.5 h), and stained with Schiff’s reagent (pararosaniline). After 1-h staining, roots were rinsed in SO2–water (three times) and then in distilled water. Root tips (1.5 mm long) were cut off and squashed in a drop of 45 % acetic acid onto slides using the dry ice method. After removing cover slips, slides were plunged into 70 % ethanol, air dried, and mounted in Canada balsam. Nuclear DNA content was evaluated by means of microdensitometry using a Jenamed 2 microscope (Carl Zeiss, Jena, Germany) with the computer-aided Cytophotometer v1.2 (Forel, Lodz, Poland). Feulgen-stained cell nuclei were measured at 565 nm.
Chemical labeling and detection of RNA transcripts
Click-iT® RNA Alexa Fluor® 488 Imaging Kit was used for visualization of RNA transcripts (Jao and Salic 2008). Root fragments (1.5 cm) were cut off and transferred to 2 mM water solution of 5-ethynyluridine (5-EU; control) or to the mixture of EU and HU, in dark. After 1.5-h labeling, 1.5-mm apical root parts were fixed in phosphate-buffered saline (PBS)-buffered 4 % paraformaldehyde (4°C; pH 7.4) for 45 min. For maceration, meristems were rinsed twice in PBS and transferred for 45 min to the citrate-buffered mixture (pH 5.0; 40°C) containing 2.5 % pectinase from A. niger, 2.5 % cellulose Onozuka R-10 from T. viride, and 2.5 % pectolyase Y-23. Next, the root tips were rinsed twice in cold PBS, squashed onto microscope slides (Polysine™, Menzel-Gläser) in a drop of distilled water, and placed on dry ice. After 10 min, cover slips were removed, and slides were washed with PBS, distilled water, and air dried. Then, the macerated cells were permeabilized with 0.5 % Triton X-100 for 15 min. Sites of EU incorporation were detected using Click-iT® reaction cocktail consisting of components prepared according to the vendor’s manual. Incubation was performed at room temperature for 60 min. After that time, slides were washed in Click-iT® reaction rinse buffer and PBS. Cell nuclei were stained with DAPI (15 μM) for 15 min and then washed in PBS. Specimens mounted in PBS/glycerol mixture (9:1) containing 2.5 % DABCO were photographed under the Eclipse E600W microscope. For Alexa Fluor® 488 DM 505 filter (excitation wavelength, 465–495 nm) and for DAPI DM 400 filter (excitation wavelength, 340–380 nm) were used.
Immunodetection of POLR2A and histone H4 Lys5 acetylation
Roots were fixed in PBS-buffered 4 % paraformaldehyde (4°C; pH 7.4) for 45 min (maceration performed as for detection of RNA transcripts) or for 20 min (isolation of cell nuclei). For isolation of nuclei, fixed meristems were rinsed twice in PBS and squashed between two slides in a drop of PBS. Fractions of isolated nuclei were harvested on microscope slides and air dried.
Immunodetection of acetylated Lys5 was performed using macerated root tips, while large subunit of RNA polymerase II (POLR2A) was visualized in fractions of isolated cell nuclei. The specimens were preincubated in the blocking buffer [5 % bovine serum albumin (BSA), 0,3 % Triton X-100, PBS] and then incubated in primary anti-acetyl-histone H4 (Lys5) antibodies (1:400, Cell Signaling) dissolved in antibody dilution buffer (1 % BSA, 0,3 % Triton X-100, PBS), or in anti-C-terminal POLR2A antibodies (1:75, Sigma) dissolved in the antibody dilution buffer (1 % BSA, PBS), in both cases overnight at 4°C. After that, slides were washed in PBS and then incubated with secondary FITC-conjugated anti-rabbit IgG (whole molecule; 1:350, Sigma) dissolved in the antibody dilution buffer (1 % BSA, 0.3 % Triton X-100, PBS), at room temperature for 90 min. Nuclear DNA was stained with DAPI (15 μM) for 15 min and then washed in PBS. The specimens were mounted and observed as described previously.
Western blotting
P-PER Plant Protein Extraction Kit supplemented with Protease Inhibitor Cocktail was used for total protein extraction. In turn, the method presented by Busk and Pages (1997) was applied to obtain nuclear proteins lysates. Whole-cell protein extracts was fractionated on NuPAGE® Novex® 4–12 % Bis-Tris gel and nuclear protein lysates on NuPAGE® Novex® 3–8 % Tris-Acetate gel and then both blotted onto polyvinylidene fluoride membrane (0.2-μm pore size). POLR2A was detected with the rabbit polyclonal anti-C-terminal domain antibodies diluted to 1:750 and goat anti-rabbit IgG antibody conjugated with alkaline phosphatase, using the Chromogenic Western Blot Immunodetection Kit.
Fluorescence intensity measurements and statistical analysis
Quantitative measurements of fluorescence were based on microscopic photographs, converted to grayscale. Computer-aided Cytophotometer v1.2 (Forel, Lodz, Poland) was used for microdensitometry analysis of fluorescence intensity. Cells were assigned to successive stage of interphase on a basis of fluorescence intensity obtained from DAPI-stained nuclei. Median values of fluorescence intensity following EU incorporation and H4 Lys5 immunodetection were estimated basing on 150 to 250 individual measurements of nuclei for each phase of the cell cycle, both in the control and HU-treated root meristems. In turn, median intensities of POLR2A immunofluorescence of nuclear regions were evaluated following 40–60 individual measurements per experimental series.
Statistical analysis was performed using Statistica 9.1 PL software. Differences between groups were assessed by Kruskal–Wallis test. A p value smaller than 0.05 was considered as statistically significant.
Results and discussion
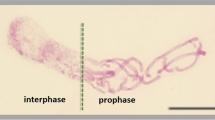
Hydroxyurea triggers the G1/S phase cell cycle arrest in root meristems of V. faba
Cytophotometric studies showed that in root meristems of V. faba treated with 2.5 mM HU for 24 h, cells accumulated preferentially in G1- and S phase (Fig. 1a, b). Similar results were obtained by Dolezel et al. (1999) following 18-h incubation in 2.5 mM HU. These data seem to be also consistent with those indicating G1/S phase arrest following HU treatment in animals (Borel et al. 2002; Lentini et al. 2006; Kaida et al. 2011) and in some plants, such as Arabidopsis thaliana (Culligan et al. 2004) or Datura inoxia (Conia et al. 1990). However, Rybaczek et al. (2008) revealed G2 arrest in root meristems of V. faba treated for 24 h with 2.5 mM HU. It should be taken into account that lots of factors may have impact on induction of phase-specific cell cycle arrest, especially when one considers that HU may not completely block replication and the cell cycle can still move forward. Occurrence of micronuclei indicated that some cells still continued cell cycle progression and preserved the ability to enter aberrant mitotic division in spite of blocked or slowed down DNA replication (data not shown). Moreover, cells of Allium cepa blocked by an intra-S checkpoint, activated in response to HU, were able to complete both their DNA synthesis and post-replication repair (Pelayo et al. 2003).
Hydroxyurea brings about changes in the dynamics of transcription and RNA polymerase II content
To evaluate the intensity of transcription in root meristem cells of V. faba, a new technique based on 5-ethynyluridine (EU) incorporation was applied. This method allows to detect newly synthesized RNA molecules by cycloaddition reaction, without applying immunocytochemical procedures. Quantitative results are shown as median fluorescence intensity. Although fluorescence measurements do not allow for drawing conclusions concerning the absolute levels of transcripts or proteins (POLR2A and H3 histone acetylated at Lys5), standardized methods and appropriate acquisition of fluorescent data let us to make comparative analyses between the two experimental series (control vs. HU). The obtained data indicate that under normal conditions, most of G1-phase cells display low fluorescence in the nucleoplasmic region. However, the number of cells with higher fluorescence increased in S- and G2 phase, and the intensity of fluorescence was enhanced 1.3-fold and 1.5-fold, respectively (Fig. 2). As compared with the control plants, HU-treated roots revealed the appearance of cells with significantly higher level of fluorescence intensity in every stage of interphase. Estimated values of fluorescence increased by average 1.8-fold in G1 phase, 1.9-fold in S phase, and 1.6-fold in G2 phase (Fig. 2).
Median fluorescence intensity (a.u. arbitrary units) in the nucleoplasmic region evaluated following 5-EU incorporation into root tip cells from seedlings incubated in H2O and HU; successive phases of the cell cycle in the control plants denoted as Control-G1, Control-S, and Control-G2 while in seedlings incubated in 2.5 mM HU (24 h) denoted as HU-G1, HU-S, and HU-G2. Statistical significance (Kruskal–Wallis test): *p < 0.001 control-G1/HU-G1, control-S/HU-S, control-G2/HU-G2; # p < 0.001 control-G1/control-S, control-G1/control-G2, HU-G1/HU-S, HU-G1/HU-G2; ^ p < 0.001 control-S/control-G2
Under normal conditions, fluorescence in nucleoli remained constant throughout all stages of the cell cycle. However, in comparison with G1- and S phases, slight increase in the fluorescence intensity has appeared in the G2-phase cells (Fig. 3). In turn, the presence of HU enhanced the number of cells displaying higher fluorescence level. Median fluorescence intensity increased 3.7-fold in G1 phase, 3.5-fold in S phase, and 2.6-fold in G2 phase, in comparison with the control (Fig. 3). The observed changes in fluorescence clearly revealed an intensified transcription following HU treatment, both in the nucleoplasmic and nucleolar regions at every phase of the cell cycle (Figs. 4 and 5).
Median fluorescence intensity (a.u. arbitrary units) in the nucleoli evaluated following 5-EU incorporation into root tip cells from seedlings incubated in H2O and HU; successive phases of the cell cycle in the control plants denoted as Control-G1, Control-S, and Control-G2 while in seedlings incubated in 2.5 mM HU (24 h) denoted as HU-G1, HU-S, and HU-G2. Statistical significance (Kruskal–Wallis test): *p < 0.001 control-G1/HU-G1, control-S/HU-S, control-G2/HU-G2; # p < 0.001 control-G1/control-G2; ^ p < 0.001 control-S/control-G2
Global character of transcription activation poses a question whether this process is accompanied by changes in RNA polymerase content or polymerase activity. High conservation within the C-terminal domains among human and plants enabled immunocytochemical identification of this protein by means of polyclonal antibodies against carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (POLR2A). Since immunofluorescence signal was restricted to euchromatin regions (heterochromatin and nucleoli were free of labeling), it can be assumed that the antibodies recognized the regions abundant in RNA polymerase II (Fig. 6). The specificity of antibodies has been tested using Western blot assay (Fig. 7). Following whole-cell protein extraction, a single band at 188 kDa was accompanied by a smear of lower molecular mass products (Fig. 7a). By using another procedure that allows for isolation of nuclear proteins, a single band has been found unexpectedly at a position corresponding to about 45 kDa (together with a fainter smear of higher mass products; Fig. 7b). Both Western blots suggest partial degradation of original POLR2A molecules and detected a 45-kDa peptide seems to be an equivalent of a CTD fragment derived from large subunit of RNA polymerase II (Kaneko and Manley 2005). Comparison between the lanes representing nuclear proteins extracted from the control and HU-treated roots indicates a slight increase in the 45-kDa peptide under stress conditions. Interestingly, previous papers reported POLR2A proteolysis during the procedure of protein isolation and consequently various electrophoretical mobility of peptides recognized by anti-POLR2A (CTD) antibodies (Guilfoyle et al. 1984; Armaleo and Gross 1985). Moreover, they point out an important problem concerning protein stability during preparation of extracts for Western blot analysis. Thus, improper conditions (e.g., pH of buffers) during isolation of some proteins may have an impact on the final interpretation of antibodies’ immunoreactivity.
Immunoblotting analysis of POLR2A in whole-cell extracts (a) and nuclear protein lysates (b). Molecular weight marker (kilodalton; lane A), electrophoresis and Coomassie staining (lane B), extracts from control seedlings (lane C) and HU-treated seedlings (lane D) immunoblotted with anti-POLR2A (CTD) antibodies
The differences in immunoblotted protein level between the two experimental series have stimulated us to perform comparative immunofluorescence analyses at the cellular level. Quantitative analysis (Fig. 8) showed changes in POLR2A content during successive phases of the cell cycle under normal conditions (1.3-fold increase of fluorescence in S phase and 2.0-fold in G2 phase, in comparison with G1 phase). However, in comparison with the control, incubation of seedlings with HU triggered a 2.3-fold increase in median fluorescence intensity in G1 phase, 2.0-fold in S phase, and 1.4-fold in G2 phase. The obtained results clearly indicate that transcription activation following HU treatment is correlated with the transport of RNA polymerase II large subunit into cell nuclei.
Median immunofluorescence intensity (a.u. arbitrary units) evaluated using anti-large subunit of RNA polymerase II (POLR2A) antibodies in seedlings incubated in H2O and HU. Successive phases of the cell cycle in the control plants denoted as Control-G1, Control-S, and Control-G2, while in seedlings incubated in 2.5 mM HU (24 h) denoted as HU-G1, HU-S, and HU-G2. Statistical significance (Kruskal–Wallis test): *p < 0.001 control-G1/HU-G1, *p < 0.001 control-S/HU-S; # p < 0.001 control-G1/control-G2; ^ p = 0.04 control-S/control-G2
Accumulation of POLR2A and enhancement of transcription in root meristems of V. faba seem to be consistent with some data indicating enhanced gene expression in response to HU treatment. Changes in transcription dynamics induced by HU did not concern stress-activated genes only (e.g., enhanced c-Fos expression; Yan and Hales 2005) but also comprise: (a) an increase in human γ-globin mRNA level (Watanapokasin et al. 2006), (b) expression of adult β-globin gene in HeLA cells (Zhang et al. 2001), (c) enhanced SMN2 (survival motor neuron) gene expression in lymphoblastoid cell lines (Grzeschik et al. 2005), and (d) accumulation of cyclin B-like proteins in A. cepa (Żabka et al. 2011). Moreover, other genotoxic stresses (such as ionizing radiation) triggered multiple gene activation in Caenorhabditis elegans (Greiss et al. 2008). However, since the majority of genes are not related to DNA damage checkpoints and DNA repair, the above authors suggest that most of them might be related to general stress responses. The obtained results are in contrary to those presented by Cui et al. (2010), who revealed transcription suppression in response to HU treatment of mouse embryonic stem cells. Taking into account the global character of transcription activation comprising both nucleoli and nucleoplasmic regions in root meristem cells of V. faba, as well as other data, it seems reasonable to conclude that not only stress response-specific genes might be activated in V. faba following HU treatment. However, this hypothesis still needs to be proved.
Hydroxyurea triggers changes in histone H4 acetylation
Since HU treatment led to transcription activation and to an increase in POLR2A content in cell nuclei of V. faba, one could ask whether this response is correlated with posttranslational modifications of histones that play a crucial role in opening up chromatin. To resolve this problem, we have concentrated on immunodetection of H4 histone acetylated at Lys5 (Fig. 9), which in V. faba was previously performed by Belyaev et al. (1997) and Jasencakova et al. (2000). Under normal conditions, the number of cells displaying intense fluorescence rose during cell cycle progression (in comparison with G1 phase, level of immunofluorescence increased 1.7- and 1.9-fold in S- and G2 phase, respectively; Fig. 10). These data are consistent with earlier reports indicating histone H4 Lys5 acetylation in TALO8 minichromosome system throughout S and G2/M phases (Unnikrishnan et al. 2010) and prior deposition of histone H4 onto DNA during replication (Sobel et al. 1995; Chang et al. 1997; Benson et al. 2006). Furthermore, HU treatment of primary roots resulted in the appearance of cells with higher fluorescence in every phase of cell cycle. In comparison with the control plants, median fluorescence intensity increased 1.9-fold in G1 phase, 1.4-fold in S phase, and 1.4-fold in G2 phase (Fig. 10).
Median immunofluorescence intensity (a.u. arbitrary units) evaluated following detection of histone H4 acetylated Lys5 in seedlings incubated in H2O and HU. Successive phases of the cell cycle in the control plants denoted as Control-G1, Control-S, and Control-G2, while in seedlings incubated in 2.5 mM HU (24 h) denoted as HU-G1, HU-S, and HU-G2. Statistical significance (Kruskal–Wallis test): *p < 0.001 control-G1/HU-G1, control-S/HU-S, control-G2/HU-G2; # p < 0.001 control-G1/control-S, control-G1/control-G2, HU-G1/HU-S, HU-G1/HU-G2
A high level of H4 Lys5 acetylation in HU-treated seedlings indicates that global transcription activation depends on histone modifications. This result seems to be consistent with that presented by Sharma et al. (2007), who showed an increase in H4 acetylation and recruitment of RNA polymerase II to RNR3 and HUG1 promoters in response to DNA-damaging agent, methyl methanesulfonate. Moreover, in Saccharomyces cerevisiae, enhanced Lys5 acetylation can be found in soluble fraction of histone H4 following HU treatment (Poveda and Sendra 2008) and at sites of DSB (Tamburini and Tyler 2005). However, McCaffrey et al. (1997) showed that HU does not affect histone acetylation in human K562 cells. In turn, Jasencakova et al. (2000) link H4 Lys5 acetylation in V. faba with replication rather than transcription. Although these data seem to be in contrast to the results presented in our paper, Belyaev et al. (1997) clearly show that unlike euchromatin, late-replicating heterochromatic regions of V. faba metaphase chromosomes are hypoacetylated, and only roots treated with trichostatin A (deacetylase inhibitor) display acetylation of heterochromatic regions in metaphase chromosomes. Therefore, obtained data still let us to assume that observed acetylation is associated with transcription, at least in the G1- and G2 phases. Furthermore, taking into account previous results of Sobel et al. (1995) and Benson et al. (2006) and those presented in our paper, it cannot be ruled out that in plants H4 Lys5 acetylation plays a role both in transcription and replication. Thus, it is reasonable to assume that acetylation observed in euchromatin during S phase may determine regions competent both for gene expression and ongoing nucleosome assembly throughout DNA synthesis.
Conclusions
Taking into account the presented results, one may ask which cellular pathways modulate transcription activation in the cells exposed to HU. Some hypotheses relevant to this problem have appeared after analyses performed using animal models. HU generates reactive oxygen species (ROS), e.g., hydrogen peroxide and nitric oxide, which induce activation of diverse genes (Sakano et al. 2001). Enhanced transcription might be triggered by mitogen-activated protein (MAP) kinase signaling pathway since ROS were found capable to activate p38 kinase in animals (Huwiler and Pfeilschifter 1999; Jia et al. 2007). However, recent analyses have shown that an increase in H4K5 acetylation following HU treatment of V. faba was reduced by applying caffeine, an inhibitor of ATM/ATR kinases (Winnicki and Maszewski, in preparation). This may indicate that both ROS and the presence of DNA lesions could directly trigger changes in transcription dynamics. In plants, this kind of ATM/ATR-mediated transcription activation in response to DSBs might also involve MAP kinase pathway since in animals following cisplatin-, doxorubicin-, or camptothecin-induced DNA damages, ATM/ATR activate p38MAPK/MK2 (Reinhardt et al. 2007). Moreover, p38 kinase was reported to be activated following HU treatment and, together with Chk1, responsible for the block of mitotic entry (Rodríguez-Bravo et al. 2007). It seems thus that a more detailed examination of the activated genes and the potential mechanisms underlying response to HU still needs to be done, especially when considering HU as a therapeutic agent.
References
Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15:2177–2196
Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK (2007) Replication in hydroxyurea: it’s a matter of time. Mol Cell Biol 27:6396–6406
Armaleo D, Gross SR (1985) Purification of the three nuclear RNA polymerases from Neurospora crassa. J Biol Chem 260:16169–16173
Bartek J, Lukas J (2001) Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 13:738–747
Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW (2002) Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A 99:8173–8178
Belyaev ND, Houben A, Baranczewski P, Schubert I (1997) Histone H4 acetylation in plant heterochromatin is altered during the cell cycle. Chromosoma 106:193–197
Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT (2006) Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem 281:9287–9296
Borel F, Lacroix FB, Margolis RL (2002) Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J Cell Sci 115:2829–2838
Busk PK, Pages M (1997) Microextraction of nuclear proteins from single maize embryos. Plant Mol Biol Rep 15:371–376
Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT (1997) Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469–480
Conia J, Alexander RG, Wilder ME, Richards KR, Rice ME, Jackson PJ (1990) Reversible accumulation of plant suspension cell cultures in G1 phase and subsequent synchronous traverse of the cell cycle. Plant Physiol 94:1568–1574
Cui P, Lin Q, Xin C, Han L, An L, Wang Y, Hu Z, Ding F, Zhang L, Hu S, Hang H, Yu J (2010) Hydroxyurea-induced global transcriptional suppression in mouse ES cells. Carcinogenesis 31:1661–1668
Culligan K, Tissier A, Britt A (2004) ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16:1091–1104
Dolezel J, Cíhalíková J, Weiserová J, Lucretti S (1999) Cell cycle synchronization in plant root meristems. Methods Cell Sci 21:95–107
Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A (2008) Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics 9:334. doi:10.1186/1471-2164-9-334
Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH (2005) Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol 58:194–202
Guilfoyle TJ, Hagen G, Malcolm S (1984) Size heterogeneity of the largest subunit of nuclear RNA polymerase II. An immunological analysis. J Biol Chem 259:649–653
Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC (2000) Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102:721–729
Huwiler A, Pfeilschifter J (1999) Nitric oxide stimulates the stress-activated protein kinase p38 in rat renal mesangial cells. J Exp Biol 202:655–660
Jackson SP (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687–696
Jao CY, Salic A (2008) Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105:15779–15784
Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I (2000) Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087–2100
Jia YT, Wei W, Ma B, Xu Y, Liu WJ, Wang Y, Lv KY, Tang HT, Wei D, Xia ZF (2007) Activation of p38 MAPK by reactive oxygen species is essential in a rat model of stress-induced gastric mucosal injury. J Immunol 179:7808–7819
Kaida A, Sawai N, Sakaguchi K, Miura M (2011) Fluorescence kinetics in HeLa cells after treatment with cell cycle arrest inducers visualized with Fucci (fluorescent ubiquitination-based cell cycle indicator). Cell Biol Int 35:359–363
Kaneko S, Manley JL (2005) The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol Cell 20:91–103
Koç A, Wheeler LJ, Mathews CK, Merrill GF (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279:223–230
Lentini L, Iovino F, Amato A, Di Leonardo A (2006) Centrosome amplification induced by hydroxyurea leads to aneuploidy in pRB deficient human and mouse fibroblasts. Cancer Lett 238:153–160
McCaffrey PG, Newsome DA, Fibach E, Yoshida M, Su MS (1997) Induction of gamma-globin by histone deacetylase inhibitors. Blood 90:2075–2083
McNeely S, Conti C, Sheikh T, Patel H, Zabludoff S, Pommier Y, Schwartz G, Tse A (2010) Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle 9:995–1004
Pelayo HR, Pincheira J, Giménez-Abián JF, Clarke DJ, de la Torre C (2003) p53-independent checkpoint controls in a plant cell model. Biol Res 36:381–388
Poveda A, Sendra R (2008) Site specificity of yeast histone acetyltransferase B complex in vivo. FEBS J 275:2122–2136
Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB (2007) p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11:175–189
Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH (2007) ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS One 2(5):e430. doi:10.1371/journal.pone.0000430
Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME (2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol 151:461–471
Rodríguez-Bravo V, Guaita-Esteruelas S, Salvador N, Bachs O, Agell N (2007) Different S/M checkpoint responses of tumor and non tumor cell lines to DNA replication inhibition. Cancer Res 67:11648–11656
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868
Rybaczek D, Kowalewicz-Kulbat M (2011) Premature chromosome condensation induced by caffeine, 2-aminopurine, staurosporine and sodium metavanadate in S-phase arrested HeLa cells is associated with a decrease in Chk1 phosphorylation, formation of phospho-H2AX and minor cytoskeletal rearrangements. Histochem Cell Biol 135:263–280
Rybaczek D, Zabka A, Pastucha A, Maszewki J (2008) Various chemical agents can induce premature chromosome condensation in Vicia faba. Acta Physiol Plant 30:663–672
Sakano K, Oikawa S, Hasegawa K, Kawanishi S (2001) Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide. Jpn J Cancer Res 92:1166–1174
Shackelford RE, Kaufmann WK, Paules RS (1999) Cell cycle control, checkpoint mechanisms, and genotoxic stress. Environ Health Perspect 107:5–24
Sharma VM, Tomar RS, Dempsey AE, Reese JC (2007) Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol 27:3199–3210
Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD (1995) Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A 92:1237–1241
Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599–602
Tamburini BA, Tyler JK (2005) Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25:4903–4913
Unnikrishnan A, Gafken PR, Tsukiyama T (2010) Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17:430–437
Watanapokasin R, Sanmund D, Winichagoon P, Muta K, Fucharoen S (2006) Hydroxyurea responses and fetal hemoglobin induction in beta-thalassemia/HbE patients’ peripheral blood erythroid cell culture. Ann Hematol 85:164–169
Yan J, Hales BF (2005) Activator protein-1 (AP-1) DNA binding activity is induced by hydroxyurea in organogenesis stage mouse embryos. Toxicol Sci 85:1013–1023
Yang J, Xu ZP, Huang Y, Hamrick HE, Duerksen-Hughes PJ, Yu YN (2004) ATM and ATR: sensing DNA damage. World J Gastroenterol 10:155–160
Żabka A, Polit JT, Maszewski J (2011) Inter- and intrachromosomal asynchrony of cell division cycle events in root meristem cells of Allium cepa: possible connection with gradient of cyclin B-like proteins. Plant Cell Rep 29:845–856
Zhang SB, He QY, Zhao H, Gui CY, Jiang C, Qian RL (2001) Function of GATA transcription factors in hydroxyurea-induced HEL cells. Cell Res 11:301–310
Acknowledgments
We thank Karolina Matczak for determining the total protein concentration in nuclear microextracts. This work was partially supported by a grant from the National Science Centre, no. 2011/01/N/NZ3/00146.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Pavla Binarova
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Winnicki, K., Polit, J.T. & Maszewski, J. Increased transcription in hydroxyurea-treated root meristem cells of Vicia faba . Protoplasma 250, 251–259 (2013). https://doi.org/10.1007/s00709-012-0402-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-012-0402-x