Abstract
Viruses of four families of arthropod-specific, large dsDNA viruses (the nuclear arthropod large DNA viruses, or NALDVs) possess homologs of genes encoding conserved components involved in the baculovirus primary infection mechanism. The presence of such homologs encoding per os infectivity factors (pif genes), along with their absence from other viruses and the occurrence of other shared characteristics, suggests a common origin for the viruses of these families. Therefore, the class Naldaviricetes was recently established, accommodating these four families. In addition, within this class, the ICTV approved the creation of the order Lefavirales for three of these families, whose members carry homologs of the baculovirus genes that code for components of the viral RNA polymerase, which is responsible for late gene expression. We further established a system for the binomial naming of all virus species in the order Lefavirales, in accordance with a decision by the ICTV in 2019 to move towards a standardized nomenclature for all virus species. The binomial species names for members of the order Lefavirales consist of the name of the genus to which the species belongs (e.g., Alphabaculovirus), followed by a single epithet that refers to the host species from which the virus was originally isolated. The common names of viruses and the abbreviations thereof will not change, as the format of virus names lies outside the remit of the ICTV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arthropod-infecting large DNA viruses belonging to four families – Baculoviridae, Nudiviridae, Hytrosaviridae, and Nimaviridae – share a set of common features that separate them from other arthropod-infecting large dsDNA viruses (Table 1). These viruses have collectively been referred to as nuclear arthropod large DNA viruses (NALDVs) [31,32,33] to distinguish them from what was previously referred to as the nucleo-cytoplasmic large DNA viruses (NCLDVs; now in the phylum Nucleocytoviricota) [9, 35]. In this paper, we explain recent developments in the classification of the NALDVs in the class Naldaviricetes and the recently established binomial nomenclature for viruses in the order Lefavirales, accommodating three of these virus families.
The pif genes as a signature for the new class Naldaviricetes
NALDVs contain genes that encode proteins collectively known as per os infectivity factors (abbreviated as PIFs; Table 2). The pif genes were originally discovered in the genomes of baculoviruses, in which they are required exclusively for oral infectivity in host insects [7, 29]. Sequencing and analysis of nudivirid, hytrosavirid, and nimavirid genomes showed that homologs of four pif genes (pif0/p74, pif1, pif2, and pif3) were conserved in the genome sequences of all of these viruses [1, 30]. More recently, a fifth pif gene (pif5/odve56) was added to this list of conserved genes [15]. Furthermore, PIF proteins are also found in the viriform particles formed in the calyx of female braconid wasps, which are classified in the genus Bracoviriform in the recently renamed family Polydnaviriformidae [5, 6, 23]. These expressed bracoviriform PIF proteins originate from the endogenization of an ancient nudivirus [5, 12, 27].
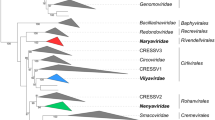
Homologs of pif genes have not been identified in other viruses and thus are signature genes for members of the NALDV families. The conservation of pif genes in NALDVs, along with other shared genomic and phenotypic characteristics, such as their rod-shaped nucleocapsids and large double-stranded circular DNA genomes that replicate in the nucleus of infected cells (Table 1), indicate a common evolutionary origin for these viruses. Consequently, it was suggested to create a higher taxon in which to classify the four NALDV families [31,32,33] as well as unassigned viruses that may share these features. This idea was further supported by the fact that phylogenetic analysis indicated that the NALDVs formed a monophyletic group, separate from the nucleocytoviricots [35]. Bipartite network analysis of dsDNA virus genes and genomes showed that NALDV genomes and the encoded core genes formed a well-supported supermodule, separate from other observed modules [13]. However, several other analytical methods have not grouped the NALDV families together [2, 36, 37], suggesting that a significant degree of genetic divergence exists between members of different NALDV families. Members of the three families in the order Herpesvirales also exhibit a high degree of genetic divergence [21], but these families are classified in the same order on the basis of shared virion structural features that allude to their common evolutionary origin [10]. Unlike the capsids of herpesvirals, the structure of the rod-like NALDV particles varies from family to family, with observable differences in dimensions (length and width), features (presence or absence of a tail and/or terminal nucleocapsid cap), and protein composition. Furthermore, sequencing of Apis mellifera filamentous virus (AmFV; currently unclassified) revealed that its genome contained homologs of the same five pif genes found in viruses of the other NALDV families [11], suggesting that this unclassified virus is also an NALDV. In sharp contrast to the rod-shaped capsids of the other NALDVs, the capsid of AmFV is a very long (> 3 µm), flexuous filament that is coiled into an envelope [3]. This observation illustrates that, in addition to genetic divergence, pif-homolog-containing large dsDNA viruses can exhibit a considerable degree of structural divergence. Also, Leptopilina boulardi filamentous virus (LbFV; Fig. 1) is currently unclassified but encodes homologs of the pif genes [20].
Phylogenetic analysis of members of the class Naldaviricetes. Concatenated alignments of five PIF amino acid sequences (pif-0/p74, pif-1, pif-2, pif-3, and pif-5/odv-e56), DNA polymerase (dnapol), and sulfhydryl oxidase (p33) were used to infer relationships by maximum likelihood as implemented in RAxML version 8.2.9 with substitution models and parameters selected for each alignment. Family-level classification is indicated for different clades in the midpoint-rooted tree. Abbreviations of virus names are as follows: AcMNPV, Autographa californica multiple nucleopolyhedrovirus; LdMNPV, Lymantria dispar MNPV; CpGV, Cydia pomonella granulovirus; CuniNPV, Culex nigripalpus nucleopolyhedrovirus; NeleNPV, Neodiprion lecontei NPV; OrNV, Oryctes rhinoceros nudivirus; GbNV, Gryllus bimaculatus NV; HzNV-2, Heliothis zea NV-2; PmNV, Penaeus monodon NV; ToNV, Tipula oleracea NV; GpSGHV, Glossina pallidipes salivary gland hypertrophy virus; MdSGHV, Musca domestica SGHV; AmFV, Apis mellifera filamentous virus; LbFV, Leptopilina boulardi FV; WSSV, white spot syndrome virus; CoBV, Chionoecetes opilio bacilliform virus. Endogenized nimaviruses from Marsupenaeus japonicus, Peneaus monodon, and Metapeneaus ensis were also included. This figure was reproduced and slightly modified from Kawato et al., 2019, J Virol 93:e01144-18, https://doi.org/10.1128/JVI.01144-18, with permission from the authors and the American Society for Microbiology.
Recently, the ICTV approved the use of taxa above the rank of “order” for virus classification [26]. We took advantage of the newly introduced hierarchy for virus classification and in 2020 proposed a class to harbour the four NALDV families (Fig. 1) (https://ictv.global/ictv/proposals/2020.006D.R.Naldaviricetes.zip). We felt that this higher rank would allow for classification of families of arthropod-infecting large dsDNA viruses that are distinguished by the inheritance of pif gene homologs but otherwise exhibit considerable genetic and structural variability and would therefore not be adequately classified within a single order. Based on the abbreviation NALDV, the approved class is named Naldaviricetes. The unclassified filamentous viruses (AmFV, LbFV) also appear to belong to this class but await assignment to species. Below, we describe why several other large dsDNA viruses that share some characteristics with the viruses now classified as Naldaviricetes members are not included in this new class.
Relationships to other taxa
Polydnaviriformidae
As indicated above, members of the genus Bracoviriform of the family Polydnaviriformidae, which infect arthropods, evolved from an ancient nudivirus that integrated its genome into the genomic DNA of an ancient parasitoid wasp [5]. The integrated nudivirus sequences in the wasp genome have retained and express pif genes in female calyx cells [8, 34]. However, the family Polydnaviriformidae also contains the genus Ichnoviriform, whose members evolved from the genome of a different, unidentified virus that integrated into parasitoid wasps of a different family [4, 19, 28]. Ichnoviriforms do not contain pif homologs and thus do not meet the criteria for classification in the proposed order Naldaviricetes. A future revision of the family Polydnaviriformidae would be needed to enable movement of the bracoviriforms to a taxon within the class Naldaviricetes, together with the Nudiviridae.
Entomopoxvirinae, Betairidovirinae and Ascoviridae
Members of these three (sub)families of large dsDNA viruses infect arthropods but lack pif homologs and have other features that distinguish them from viruses in the class Naldaviricetes (Table 1) [37]. Entomopoxvirins and betairidovirins possess linear genomes, which are partially or wholly synthesized in the cytoplasm of infected cells. Ascovirids have circular genomes whose replication is initiated in the nucleus, but they clearly share a more recent origin with viruses in the subfamily Betairidovirinae [24]. In 2020, the ICTV ratified a taxonomic proposal to create the order Pimascovirales for the families Ascoviridae, Iridoviridae, and Marseilleviridae [17]. The proposal also created a realm, Varidnaviria, in which the families of the order Pimascovirales together with the other nucleocytoviricots are classified. The distinguishing characteristic of viruses classified in the realm Varidnaviria is the occurrence of a virus hallmark gene encoding a vertical double jelly-roll major capsid protein (VDJ-MCP). The members of the Naldaviricetes, on the other hand, do not contain homologs encoding a VDJ-MCP, but they do have other “connector” genes that might link the “baculo-like” supermodule with the nucleocytoviricot-polinton supermodule in dsDNA virus gene/genome networks [13]. This observation suggests that naldoviricetes may form an ancient branch of the Varidnaviria that has evolved to use different proteins for capsid assembly [13, 16]. However, we are presently not proposing to place the class Naldaviricetes into the Varidnaviria hierarchy.
Establishment of the order Lefavirales within the class Naldaviricetes
Phylogenies based on various naldaviricete sequence alignments often place viruses of the families Baculoviridae, Nudiviridae, and Hytrosaviridae into a clade separate from the Nimaviridae [6, 15, 29, 31]. Baculoviruses and viruses classified as members of the Nudiviridae and Hytrosaviridae contain homologs of genes that encode components of the baculovirus late-phase transcription complex, including three of the four subunits of the baculovirus DNA-directed RNA polymerase (lef-4, lef-8, and lef-9) (Table 2) [14, 22]. These homologs are not found in the genomes of nimavirids. We thus created an order within the class Naldaviricetes into which the families Baculoviridae, Nudiviridae, and Hytrosaviridae were placed (Fig. 2). This order is named Lefavirales, from the term “late expression factor” (abbreviated as lef), which was previously coined to refer to genes identified in a screen for ORFs required for (or supporting) late-phase baculovirus transcription [22]. Lefavirals are characterized by the possession of conserved baculovirus transcription gene homologs and can also be distinguished from nimavirids in phylogenetic analysis (see Fig. 1). At present, we have refrained from creating an order for the family Nimaviridae, as there is insufficient information from which to extrapolate the distinguishing features of viruses in such an order. This strategy is consistent with the International Code of Virus Classification and Nomenclature (ICVCN; March 2021) Rule 3.2, which indicates that it is not mandatory to use all levels of the taxonomic hierarchy.
Taxonomic hierarchy of families of nuclear arthropod large DNA viruses. A new class, Naldaviricetes, was established for classification of the viruses in the four currently established families Baculoviridae, Nudiviridae, Hytrosaviridae, and Nimaviridae. A new order, Lefavirales, was introduced to include three of these families. The two viruses indicated at the top of the figure (Apis mellifera filamentous virus [3, 10] and Leptopilina boulardi filamentous virus [15]) are currently unclassified, but based on their genome content, they bear the hallmarks of members of the taxa Naldaviricetes and Lefavirales, respectively.
Binomial naming system for virus species in the order Lefavirales
In 2021, the ICTV membership ratified a proposal (2018.001G.R) to adopt a binomial virus species naming system that follows the method of Linnaeus. This means that the Linnaean binomial format needs to be implemented for all virus species names, with a 2023 deadline. Accordingly, ICVCN Rule 3.21 now reads:
"A species name shall consist of only two distinct word components separated by a space.
The first word component shall begin with a capital letter and be identical in spelling to the name of the genus to which the species belongs. The second word component shall not contain any suffixes specific for taxa of higher ranks. The entire species name (both word components) shall be italicized”.
Since the order Lefavirales has three families, Baculoviridae, Nudiviridae, and Hytrosaviridae, it makes sense to name the species belonging to these families in a similar way. The two ICTV Study Groups concerned have therefore joined forces, designed a general method to convert all existing species names into binomial names, and submitted a formal ICTV taxonomic proposal in 2022 for consideration (https://ictv.global/ictv/proposals/2022.003D.Lefavirales_106rensp.zip). The same strategy is also used to assign species names to newly discovered lefavirals. Below, we present the new system and explain the reasoning behind the chosen method. As such, we aim to provide guidance for scientists in the field for naming new lefaviral species. In the following explanations, we will use baculovirids as examples. All updated lefaviral species names are provided in Table 3. Please be aware that the renaming only applies to virus species names. The names of viruses and their isolates remain unchanged. Since only names of virus species and higher taxa are regulated by the ICTV, it is expected that the way the viruses themselves are routinely named in the literature will remain unchanged. Thus, for naming old and new naldaviricete virus isolates, the historic conventions and practices should be continued as indicated in Table 3.
Issues with the previous lefaviral species naming system
In the past, the species names for lefavirals varied in format from family to family. For the family Baculoviridae, species names previously consisted of the binomial name of the host species, sometimes followed by a virion structural characteristic (“multiple” in Autographa californica multiple nucleopolyhedrovirus) and/or by a now obsolete genus name (nucleopolyhedrovirus, granulovirus). In most cases the virus species name did not differ from the virus name, except that the virus species name was fully written in italic letters, which made it often complicated to distinguish between the virus species and the virus itself. Nudivirid species names also started with the host species name, followed by the common name for viruses of this family (nudivirus). Species names in the family Hytrosaviridae consisted of the genus name of the host (e.g., Glossina) followed by the virus genus name (hytrosavirus). Species names for all three families featured part or all of the species names of the viral hosts. In developing specific epithets for lefaviral species names, we have retained this familiar feature in order to ease the transition to a new binomial format that is consistent among all lefaviral families.
The remaining family in the class Naldaviricetes, the Nimaviridae, was not included in this proposed binomial naming system, as the naming of the only classified species in this family is historically based on symptoms and not on host species, in contrast to lefaviral species, but it would be logical to follow the same principle for nimavirid species.
Binomial naming method for virus species in the order Lefavirales
The binomial species names for lefavirals are composed as follows:
-
As for Linnaean binomial species names in general, the first word of the species name is the name of the genus to which the virus species belongs, starting with a capital (e.g., Alphabaculovirus or Betanudivirus).
-
The second word (the epithet) reflects the Latin species name of the arthropod host from which the virus was originally isolated. It is composed of the first 2–4 letters of the host genus directly coupled to the genitive form of the epithet of the host species name. For example, Autographa californica multiple nucleopolyhedrovirus is now Alphabaculovirus aucalifornicae, and Cydia pomonella granulovirus is now Betabaculovirus cypomonellae. The specific epithet should be readable and pronounceable.
-
Latin ordinal prefixes are added to the specific epithet to distinguish between species with isolates originating from the same host. When a second species from the same host is identified, the prefix “alter-“ is placed at the start of the epithet. For example, Mamestra configurata nucleopolyhedrovirus A, the first species identified from the bertha armyworm, Mamestra configurata, is now Alphabaculovirus maconfiguratae, while Mamestra configurata nucleopolyhedrovirus B, the second species with isolates identified from M. configurata, became Alphabaculovirus altermaconfiguratae. For subsequent species to classify viruses isolated from the same host, the appropriate Latin ordinal prefixes will be added to the specific epithet. For example, “tert(i)” and “quart(u)” will be placed in front of “-maconfiguratae” if alphabaculoviruses representing a third and fourth distinct species from M. configurata would be identified.
Explanations and examples of the epithet strategy
It might seem simpler to adapt the host-specific epithet alone as the specific epithet for lefaviral species, as was done for the microsporidium Nosema ceranae, a pathogen of the Asiatic honey bee, Apis cerana. However, this approach does not account for situations in which two distinct viruses from the same genus are isolated from different hosts that share the same specific epithet. There is already an example of this situation: in addition to Autographa californica multiple nucleopolyhedrovirus (already classified as Alphabaculovirus aucalifornicae), there is another alphabaculovirus identified from the California oakworm, Phryganidia californica [18]. Adding the 2–4 first letters of the host’s genus name as the start of the epithet resolves this problem.
The genus name, per definition, ends in “-virus”, and as a consequence, the genus names are all of neuter gender in Latin. We can therefore not simply use the original epithet of the host, which may have been of female, male, or neuter gender. Therefore, we proposed to use the genitive form of the epithet of the insect species in the binomial name of the virus species. Genitive forms have the meaning: “owned by, derived from, belonging to”. (The singular genitive form of most Latin words ends in -ae, -i, or -is, depending on the declension. All plural genitive forms end in -um). For explanations of less obvious epithets, see Supplementary Table S1.
In the situation where the host epithet already appears to be neuter, there would not be a strict linguistic need to change it. However, for overall consistency, we decided to use the genitive form there as well. Example: Betabaculovirus xecnigri (from the host Xestia c-nigrum). But what to do if the host’s epithet is already in the genitive form or looks the same as a genitive form? Then we will leave it as it is, as the use of double genitives is not useful. This is exemplified by Alphabaculovirus anpernyi (from the host Antheraea pernyi), Betabaculovirus cnamedinalis (from Cnaphalocrocis medinalis), Betabaculovirus agsegetum (already genitive plural in Agrotis segetum; from “seges”, meaning from the grain fields/crops).
In the past, a capital letter was appended to the end of baculovirus species names to distinguish species with isolates originating from the same host, as described above for alphabaculoviruses from M. configurata. One simple approach to reproducing this solution in the context of a binomial system might have been to attach the letter to the specific epithet of a binomial name with a hyphen. However, the use of hyphens to attach numbers or letters to the end of a series of species names is specifically excluded by ICVCN Rule 3.13. Thus, ordinal prefixes are used to distinguish different species isolated from the same host, as described above. The exact form of the ordinal prefixes will depend on the ease of pronunciation of the resulting epithet.
The background for the adopted strategy and some more general rules for composing epithets can be found in: “Advice and guidelines to Study Groups on the implementation of binomial species names", to be found at https://ictv.global/filebrowser/download/435 or in the recently published paper by Postler and collaborators [25].
Conclusions
In this paper, we report recent major changes in the taxonomy of NALVDs, which are now part of the official ICTV taxonomy. The class Naldaviricetes and the order Lefavirales were established in 2021. The binomial species naming system for lefavirals was ratified in April 2023. In case of questions on how to name new virus species, please contact the respective ICTV Study Group. It is further proposed to continue using conventional naming and abbreviations for virus isolates, which will facilitate distinguishing between viruses and virus species.
Change history
20 September 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00705-023-05857-9
References
Abd-Alla AM, Vlak JM, Bergoin M, Maruniak JE, Parker A, Burand JP, Jehle JA, Boucias DG, Hytrosavirus Study Group of the I (2009) Hytrosaviridae: a proposal for classification and nomenclature of a new insect virus family. Arch Virol 154:909–918
Aiewsakun P, Simmonds P (2018) The genomic underpinnings of eukaryotic virus taxonomy: creating a sequence-based framework for family-level virus classification. Microbiome 6:38
Bailey L, Carpenter JM, Woods RD (1981) Properties of a filamentous virus of the honey bee (Apis mellifera). Virology 114:1–7
Beliveau C, Cohen A, Stewart D, Periquet G, Djoumad A, Kuhn L, Stoltz D, Boyle B, Volkoff AN, Herniou EA, Drezen JM, Cusson M (2015) Genomic and proteomic analyses indicate that banchine and campoplegine polydnaviruses have similar, if not identical, viral ancestors. J Virol 89:8909–8921
Bezier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M, Pfister-Wilhem R, Periquet G, Dupuy C, Huguet E, Volkoff AN, Lanzrein B, Drezen JM (2009) Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930
Bezier A, Theze J, Gavory F, Gaillard J, Poulain J, Drezen JM, Herniou EA (2015) The genome of the nucleopolyhedrosis-causing virus from Tipula oleracea sheds new light on the Nudiviridae family. J Virol 89:3008–3025
Boogaard B, van Oers MM, van Lent JWM (2018) An advanced view on baculovirus per os infectivity factors. Insects 9
Burke GR, Thomas SA, Eum JH, Strand MR (2013) Mutualistic polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathog 9:e1003348
Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D (2013) "Megavirales", a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521
Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E (2009) The order Herpesvirales. Arch Virol 154:171–177
Gauthier L, Cornman S, Hartmann U, Cousserans F, Evans JD, de Miranda JR, Neumann P (2015) The Apis mellifera filamentous virus genome. Viruses 7:3798–3815
Herniou EA, Huguet E, Theze J, Bezier A, Periquet G, Drezen JM (2013) When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos Trans R Soc Lond B Biol Sci 368:20130051
Iranzo J, Krupovic M, Koonin EV (2016) The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7
Jehle JA, Abd-Alla AM, Wang Y (2013) Phylogeny and evolution of Hytrosaviridae. J Invertebr Pathol 112 Suppl:S62–67
Kawato S, Shitara A, Wang Y, Nozaki R, Kondo H, Hirono I (2019) Crustacean genome exploration reveals the evolutionary origin of white spot syndrome virus. J Virol 93
Koonin EV, Dolja VV, Krupovic M (2015) Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 479–480:2–25
Koonin EVDV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH (2019) (2019) Create a megataxonomic framework, filling all principal taxonomic ranks, for DNA viruses encoding vertical jelly-roll type major capsid proteins. https://ictv.global/ICTV/proposals/2019.003G.zip
Lange M, Wang H, Zhihong H, Jehle JA (2004) Towards a molecular identification and classification system of lepidopteran-specific baculoviruses. Virology 325:36–47
Legeai F, Santos BF, Robin S, Bretaudeau A, Dikow RB, Lemaitre C, Jouan V, Ravallec M, Drezen JM, Tagu D, Baudat F, Gyapay G, Zhou X, Liu S, Webb BA, Brady SG, Volkoff AN (2020) Genomic architecture of endogenous ichnoviruses reveals distinct evolutionary pathways leading to virus domestication in parasitic wasps. BMC Biol 18:89
Lepetit D, Gillet B, Hughes S, Kraaijeveld K, Varaldi J (2016) Genome sequencing of the behavior manipulating virus LbFV reveals a possible new virus family. Genome Biol Evol 8:3718–3739
McGeoch DJ, Rixon FJ, Davison AJ (2006) Topics in herpesvirus genomics and evolution. Virus Res 117:90–104
Passarelli AL, Guarino LA (2007) Baculovirus late and very late gene regulation. Curr Drug Targets 8:1103–1115
Petersen JM, Bezier A, Drezen JM, van Oers MM (2022) The naked truth: An updated review on nudiviruses and their relationship to bracoviruses and baculoviruses. J Invertebr Pathol 189:107718
Piegu B, Asgari S, Bideshi D, Federici BA, Bigot Y (2015) Evolutionary relationships of iridoviruses and divergence of ascoviruses from invertebrate iridoviruses in the superfamily Megavirales. Mol Phylogenet Evol 84:44–52
Postler TS, Rubino L, Adriaenssens EM, Dutilh BE, Harrach B, Junglen S, Kropinski AM, Krupovic M, Wada J, Crane A, Kuhn JH, Mushegian A, Rumnieks J, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Callanan J, Draper LA, Hill C, Stockdale SR (2022) Guidance for creating individual and batch latinized binomial virus species names. J Gen Virol 103
Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Sanfacon H, Simmonds P, Varsani A, Zerbini FM, Davison AJ (2019) Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch Virol 164:943–946
Thezé J, Bezier A, Periquet G, Drezen JM, Herniou EA (2011) Paleozoic origin of insect large dsDNA viruses. Proc Natl Acad Sci U S A 108:15931–15935
Volkoff AN, Jouan V, Urbach S, Samain S, Bergoin M, Wincker P, Demettre E, Cousserans F, Provost B, Coulibaly F, Legeai F, Beliveau C, Cusson M, Gyapay G, Drezen JM (2010) Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog 6:e1000923
Wang X, Liu X, Makalliwa GA, Li J, Wang H, Hu Z, Wang M (2017) Per os infectivity factors: a complicated and evolutionarily conserved entry machinery of baculovirus. Sci China Life Sci 60:806–815
Wang X, Chen C, Zhang N, Chen Q, Zhang F, Liu X, Li F, Shi ZL, Vlak JM, Wang M, Hu Z (2022) Functional peroral infectivity complex of White Spot Syndrome Virus of shrimp. J Virol 96:e0117322
Wang Y, Jehle JA (2009) Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol 101:187–193
Wang Y, Bininda-Emonds OR, van Oers MM, Vlak JM, Jehle JA (2011) The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes 42:444–456
Wang Y, O.R.P. B-E, Jehle JA (2012) Nudivirus genomics and phylogeny. In: Maria Laura G, Victor R (eds) Viral Genomes. IntechOpen, Rijeka, pp 33–52
Wetterwald C, Roth T, Kaeslin M, Annaheim M, Wespi G, Heller M, Maser P, Roditi I, Pfister-Wilhelm R, Bezier A, Gyapay G, Drezen JM, Lanzrein B (2010) Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J Gen Virol 91:2610–2619
Williams T, Bergoin M, van Oers MM (2017) Diversity of large DNA viruses of invertebrates. J Invertebr Pathol 147:4–22
Wu GA, Jun SR, Sims GE, Kim SH (2009) Whole-proteome phylogeny of large dsDNA virus families by an alignment-free method. Proc Natl Acad Sci U S A 106:12826–12831
Yutin N, Koonin EV (2012) Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Robert Harrison, Elisabeth Herniou, Johannes Jehle, David Theilmann, Peter Krell, and Monique van Oers were authors of the Naldaviricetes ICTV proposal (2020.006D.R.Naldaviricetes.zip). Monique van Oers, Adly Abd-Alla, and Robert Harrison prepared the ICTV proposal for the binomial naming of Lefavirales (2022.003D.A.Lefavirales_106rensp.docx), with input from the other authors, except David Theilmann. Monique van Oers drafted the first version of this manuscript based on the two proposals mentioned here. All authors commented on previous versions of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose
Additional information
Communicated by Sead Sabanadzovic
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Legends of Fig.1 and Fig.2 have been interchanged. Table 3 corrected.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Oers, M.M., Herniou, E.A., Jehle, J.A. et al. Developments in the classification and nomenclature of arthropod-infecting large DNA viruses that contain pif genes. Arch Virol 168, 182 (2023). https://doi.org/10.1007/s00705-023-05793-8
Published:
DOI: https://doi.org/10.1007/s00705-023-05793-8