Abstract
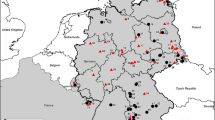
Rodents host different orthohepeviruses, namely orthohepevirus C genotype HEV-C1 (rat hepatitis E virus, HEV) and the additional putative genotypes HEV-C3 and HEV-C4. Here, we screened 2,961 rodents from Central Europe by reverse transcription polymerase chain reaction (RT-PCR) and identified HEV RNA in 13 common voles (Microtus arvalis) and one bank vole (Myodes glareolus) with detection rates of 2% (95% confidence interval [CI]: 1-3.4) and 0.08% (95% CI: 0.002-0.46), respectively. Sequencing of a 279-nucleotide RT-PCR amplicon corresponding to a region within open reading frame (ORF) 1 showed a high degree of similarity to recently described common vole-associated HEV (cvHEV) sequences from Hungary. Five novel complete cvHEV genome sequences from Central Europe showed the typical HEV genome organization with ORF1, ORF2 and ORF3 and RNA secondary structure. Uncommon features included a noncanonical start codon in ORF3, multiple insertions and deletions within ORF1 and ORF2/ORF3, and the absence of a putative ORF4. Phylogenetic analysis showed all of the novel cvHEV sequences to be monophyletic, clustering most closely with an unassigned bird-derived sequence and other sequences of the species Orthohepevirus C. The nucleotide and amino acid sequence divergence of the common vole-derived sequences was significantly correlated with the spatial distance between the trapping sites, indicating mostly local evolutionary processes. Detection of closely related HEV sequences in common voles in multiple localities over a distance of 800 kilometers suggested that common voles are infected by cvHEV across broad geographic distances. The common vole-associated HEV strain is clearly divergent from HEV sequences recently found in narrow-headed voles (Microtus gregalis) and other cricetid rodents.





Similar content being viewed by others
References
Andonov A, Robbins M, Borlang J, Cao J, Hattchete T, Stueck A, Deschaumbault Y, Murnaghan K, Varga J, Johnston B (2019) Rat hepatitis E virus linked to severe acute hepatitis in an immunocompetent patient. J Infect Dis. https://doi.org/10.1093/infdis/jiz025
Batts W, Yun S, Hedrick R, Winton J (2011) A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158:116–123
Beysard M, Heckel G (2014) Structure and dynamics of hybrid zones at different stages of speciation in the common vole (Microtus arvalis). Mol Ecol 23:673–687
Cao D, Huang YW, Meng XJ (2010) The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84:13040–13044
Clopper CJ, Pearson ES (1934) The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413
Computing RFfS (2015) R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. http://www.R-project.org
Cossaboom CM, Cordoba L, Sanford BJ, Pineyro P, Kenney SP, Dryman BA, Wang Y, Meng XJ (2012) Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J Gen Virol 93:1687–1695
de Souza WM, Romeiro MF, Sabino-Santos G Jr, Maia FGM, Fumagalli MJ, Modha S, Nunes MRT, Murcia PR, Figueiredo LTM (2018) Novel orthohepeviruses in wild rodents from Sao Paulo State, Brazil. Virology 519:12–16
Ding Q, Heller B, Capuccino JM, Song B, Nimgaonkar I, Hrebikova G, Contreras JE, Ploss A (2017) Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci USA 114:1147–1152
Dremsek P, Wenzel JJ, Johne R, Ziller M, Hofmann J, Groschup MH, Werdermann S, Mohn U, Dorn S, Motz M, Mertens M, Jilg W, Ulrich RG (2012) Seroprevalence study in forestry workers from eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med Microbiol Immunol 201:189–200
Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose SM, Adu-Sarkodie Y, Oppong SK, Kalko EK, Osterman A, Rasche A, Adam A, Muller MA, Ulrich RG, Leroy EM, Lukashev AN, Drosten C (2012) Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86:9134–9147
Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C (2013) Evidence for novel hepaciviruses in rodents. PLoS Pathog 9:e1003438
Guan D, Li W, Su J, Fang L, Takeda N, Wakita T, Li TC, Ke C (2013) Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg Infect Dis 19:1341–1343
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 41
Hjelle B, Yates T (2001) Modeling hantavirus maintenance and transmission in rodent communities. Curr Top Microbiol Immunol 256:77–90
Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV (2011) Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res 39:4220–4234
Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL (2012) Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18:1274–1281
Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich RG (2010) Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16:1452–1455
Johne R, Plenge-Bonig A, Hess M, Ulrich RG, Reetz J, Schielke A (2010) Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol 91:750–758
Johne R, Dremsek P, Reetz J, Heckel G, Hess M, Ulrich RG (2014) Hepeviridae: an expanding family of vertebrate viruses. Infect, Genet Evolut 27:212–229
Johne R, Reetz J, Ulrich RG, Machnowska P, Sachsenroder J, Nickel P, Hofmann J (2014) An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J Viral Hepat 21:447–456
Kanai Y, Miyasaka S, Uyama S, Kawami S, Kato-Mori Y, Tsujikawa M, Yunoki M, Nishiyama S, Ikuta K, Hagiwara K (2012) Hepatitis E virus in Norway rats (Rattus norvegicus) captured around a pig farm. BMC Res Notes 5:4
Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW (1992) Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci USA 89:8259–8263
Kurucz K, Hederics D, Bali D, Kemenesi G, Horvath G, Jakab F (2019) Hepatitis E virus in Common voles (Microtus arvalis) from an urban environment, Hungary: Discovery of a Cricetidae-specific genotype of Orthohepevirus C. Zoonoses Publ Health 66:259–263. https://doi.org/10.1111/zph.12543
Lack JB, Volk K, Van Den Bussche RA (2012) Hepatitis E virus genotype 3 in wild rats, United States. Emerg Infect Dis 18:1268–1273
Li TC, Ami Y, Suzaki Y, Yasuda SP, Yoshimatsu K, Arikawa J, Takeda N, Takaji W (2013) Characterization of full genome of rat hepatitis E virus strain from Vietnam. Emerg Infect Dis 19:115–118
Li W, Guan D, Su J, Takeda N, Wakita T, Li TC, Ke CW (2013) High prevalence of rat hepatitis E virus in wild rats in China. Vet Microbiol 165:275–280
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203
Martin DP, Posada D, Crandall KA, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrovir 21:98–102
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 1(1):vev003
Meerburg BG, Singleton GR, Kijlstra A (2009) Rodent-borne diseases and their risks for public health. Crit Rev Microbiol 35:221–270
Mulyanto Depamede SN, Sriasih M, Takahashi M, Nagashima S, Jirintai S, Nishizawa T, Okamoto H (2013) Frequent detection and characterization of hepatitis E virus variants in wild rats (Rattus rattus) in Indonesia. Arch Virol 158:87–96
Mulyanto Suparyatmo JB, Andayani IG, Khalid Takahashi M, Ohnishi H, Jirintai S, Nagashima S, Nishizawa T, Okamoto H (2014) Marked genomic heterogeneity of rat hepatitis E virus strains in Indonesia demonstrated on a full-length genome analysis. Virus Res 179:102–112
Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P (2017) Host and viral traits predict zoonotic spillover from mammals. Nature 546:646–650
Primadharsini PP, Mulyanto Wibawa IDN, Anggoro J, Nishizawa T, Takahashi M, Jirintai S, Okamoto H (2018) The identification and characterization of novel rat hepatitis E virus strains in Bali and Sumbawa, Indonesia. Arch Virol 163:1345–1349
Purcell RH, Engle RE, Rood MP, Kabrane-Lazizi Y, Nguyen HT, Govindarajan S, St Claire M, Emerson SU (2011) Hepatitis E virus in rats, Los Angeles, California, USA. Emerg Infect Dis 17:2216–2222
Purdy MA, Harrison TJ, Jameel S, Meng XJ, Okamoto H, Van der Poel WHM, Smith DB, ICTV Report C (2017) ICTV virus taxonomy profile: Hepeviridae. J Gen Virol 98:2645–2646
Reuter G, Boros A, Matics R, Kapusinszky B, Delwart E, Pankovics P (2016) Divergent hepatitis E virus in birds of prey, common kestrel (Falco tinnunculus) and red-footed falcon (F. vespertinus), Hungary. Infect, Genet Evolut 43:343–346
Ryll R, Bernstein S, Heuser E, Schlegel M, Dremsek P, Zumpe M, Wolf S, Pepin M, Bajomi D, Muller G, Heiberg AC, Spahr C, Lang J, Groschup MH, Ansorge H, Freise J, Guenther S, Baert K, Ruiz-Fons F, Pikula J, Knap N, Tsakmakidis I, Dovas C, Zanet S, Imholt C, Heckel G, Johne R, Ulrich RG (2017) Detection of rat hepatitis E virus in wild Norway rats (Rattus norvegicus) and Black rats (Rattus rattus) from 11 European countries. Vet Microbiol 208:58–68
Saxenhofer M, Weber de Melo V, Ulrich RG, Heckel G (2017) Revised time scales of RNA virus evolution based on spatial information. Proc R Soc B: Biol Sci 284:20170857
Saxenhofer M, Schmidt S, Ulrich RG, Heckel G (2019) Secondary contact between diverged host lineages entails ecological speciation in a European hantavirus. PLoS Biol 17:e3000142
Shimizu K, Hamaguchi S, Ngo CC, Li TC, Ando S, Yoshimatsu K, Yasuda SP, Koma T, Isozumi R, Tsuda Y, Fujita H, Pham TT, Le MQ, Dang AD, Nguyen TQ, Yoshida LM, Ariyoshi K, Arikawa J (2016) Serological evidence of infection with rodent-borne hepatitis E virus HEV-C1 or antigenically related virus in humans. J Vet Med Sci Jpn Soc Vet Sci 78:1677–1681
Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU (2012) Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86:5697–5707
Smith DB, Simmonds P, International Committee on Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA (2014) Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232
Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA (2016) Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol 97:537–542
Spahr C, Ryll R, Knauf-Witzens T, Vahlenkamp TW, Ulrich RG, Johne R (2017) Serological evidence of hepatitis E virus infection in zoo animals and identification of a rodent-borne strain in a Syrian brown bear. Vet Microbiol 212:87–92
Sridhar S, Yip CCY, Wu S, Cai J, Zhang AJ, Leung KH, Chung TWH, Chan JFW, Chan WM, Teng JLL, Au-Yeung RKH, Cheng VCC, Chen H, Lau SKP, Woo PCY, Xia NS, Lo CM, Yuen KY (2018) Rat hepatitis E virus as cause of persistent hepatitis after liver transplant. Emerg Infect Dis 24:2241–2250
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evolut 30:2725–2729
Wang B, Li W, Zhou JH, Li B, Zhang W, Yang WH, Pan H, Wang LX, Bock CT, Shi ZL, Zhang YZ, Yang XL (2018) Chevrier’s field mouse (Apodemus chevrieri) and Pere David’s vole (Eothenomys melanogaster) in China carry orthohepeviruses that form two putative novel genotypes within the species orthohepevirus C. Virol Sinica 33:44–58
Wilson DE, Reeder DM (2005) Mammal species of the world: a taxonomic and geographic reference. JHU Press, Baltimore
Wolf S, Reetz J, Johne R, Heiberg AC, Petri S, Kanig H, Ulrich RG (2013) The simultaneous occurrence of human norovirus and hepatitis E virus in a Norway rat (Rattus norvegicus). Arch Virol 158:1575–1578
Woo PC, Lau SK, Teng JL, Cao KY, Wernery U, Schountz T, Chiu TH, Tsang AK, Wong PC, Wong EY, Yuen KY (2016) New hepatitis E virus genotype in bactrian camels, Xinjiang, China, 2013. Emerg Infect Dis 22:2219–2221
Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, Jiang J, Yang J, Dong J, Sun L, Zhu Y, Li Y, Zheng D, Zhang C, Su H, Zheng Y, Zhou H, Zhu G, Li H, Chmura A, Yang F, Daszak P, Wang J, Liu Q, Jin Q (2018) Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178
Yamada K, Takahashi M, Hoshino Y, Takahashi H, Ichiyama K, Nagashima S, Tanaka T, Okamoto H (2009) ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol 90:1880–1891
Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, Shima R, Moriishi K, Tsukihara T, Li TC, Takeda N, Miyamura T, Matsuura Y (2009) Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci USA 106:12986–12991
Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y (2009) A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81:1371–1379
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgements
The authors thank Stephan Eichenberg for creating the map, and Filip Cierniak for helpful discussions.
Author information
Authors and Affiliations
Contributions
RGU and JFD designed the study; RR and JFD performed the analyses; RR, VMC, JFD, GH and RGU wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Funding
This study was funded in part by the German Federal Ministry of Health (ZMVI1-2518FSB705) to VMC, by DZIF (TTU Emerging Infections, network RaBoPa) to RGU, and by the Swiss National Science Foundation (31003A-176209) to GH.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animal experiments performed by any of the authors.
Additional information
Handling Editor: Michael A. Purdy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ryll, R., Heckel, G., Corman, V.M. et al. Genomic and spatial variability of a European common vole hepevirus. Arch Virol 164, 2671–2682 (2019). https://doi.org/10.1007/s00705-019-04347-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04347-1