Abstract
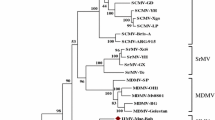
Based on serology, cytopathology, cereal host range and leafhopper vector, maize sterile stunt virus (MSSV) has been regarded as a strain of the cytorhabdovirus barley yellow striate mosaic virus (BYSMV). Here, we report the first-ever sequence of MSSV, comprising the complete genome of 12,561 nucleotides. Detailed analysis of genome organization, coding and non-coding sequences, and phylogeny confirms the close relationship to BYSMV and supports classification of this virus a strain of BYSMV.


Similar content being viewed by others
References
Greber RS (1982) Maize sterile stunt—a delphacid transmitted rhabdovirus disease affecting some maize genotypes in Australia. Aust J Agric Res 33:13–23
Walker PJ, Blasdell KR, Calisher CH, Dietzgen RG, Kondo H, Kurath G, Longdon B, Stone DM, Tesh RB, Tordo N, Vasilakis N, Whitfield AE (2018) ICTV virus taxonomy profile: Rhabdoviridae. J Gen Virol 99:447–448
Milne RG, Masenga V, Conti M (1986) Serological relationships between the nucleocapsids of some planthopper-borne rhabdoviruses of cereals. Intervirology 25:83–87
Lockhart BEL, El Maataoui M, Carroll TW, Lennon AM, Zaske SK (1986) Identification of barley yellow striate mosaic virus in Morocco and its field detection by enzyme immune assay. Plant Dis 70:1113–1117
Greber RS (1990) Discrimination by gel-diffusion serology of digitaria striate, maize sterile stunt and other rhabdoviruses of Poacea. Ann Appl Biol 116:259–264
Milne RG, Conti M (1986) Barley yellow striate mosaic virus. In: CMI/AAB descriptions of plant viruses, vol 312. Association of Applied Biologists, Kew, UK
Yan T, Zhu J-R, Di D, Gao Q, Zhang Y, Zhang A, Yan C, Miao H, Wang X-B (2015) Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478:112–122
Almasi R, Afsharifar A, Niazi A, Pakdel A, Izadpanah K (2010) Analysis of the complete nucleotide sequence of the polymerase gene of Barley yellow striate mosaic virus—Iranian isolate. J Phytopathol 158:351–356
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320
Maurino F, Dumón AD, Llauger G, Alemandri V, de Haro LA, Mattio MF, del Vas M, Laguna IG, de la Paz Giménez Pecci M (2018) Complete genome sequence of maize yellow striate virus, a new cytorhabdovirus infecting maize and wheat crops in Argentina. Arch Virol 163:291–295
Tanno F, Nakatsu A, Toriyama S, Kojima M (2000) Complete nucleotide sequence of Northern cereal mosaic virus and its genome organization. Arch Virol 145:1373–1384
Acknowledgements
We thank Denis Persley for assistance in collecting the field samples, and David Innes for 454 sequence assembly. This research was jointly supported by the Queensland Department of Agriculture and Fisheries and the University of Queensland through the Queensland Alliance for Agriculture and Food Innovation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: F. Murilo Zerbini.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dietzgen, R.G., Higgins, C.M. Complete genome sequence of maize sterile stunt virus. Arch Virol 164, 1221–1223 (2019). https://doi.org/10.1007/s00705-019-04164-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04164-6