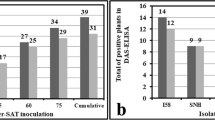
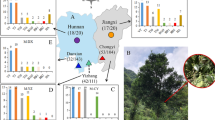
Abstract
Stem-pitting (SP) is the main type of citrus tristeza virus (CTV) that causes severe damage to citrus trees, especially those of sweet orange, in Hunan province, China. Understanding the local CTV population structure should provide clues for effective mild strain cross-protection (MSCP) of the SP strain of CTV. In this study, markers for the p23 gene, multiple molecular markers (MMMs), and sequence analysis of the three silencing suppressor genes (p20, p23 and p25) were employed to analyze the genetic diversity and genotype composition of the CTV population based on 51 CTV-positive samples collected from 14 citrus orchards scattered around six major citrus-growing areas of Hunan. The results indicated that the CTV population structure was extremely complex and that infection was highly mixed. In total, p23 gene markers resulted in six profiles, and MMMs demonstrated 25 profiles. The severe VT and T3 types appeared to be predominantly associated with SP, while the mild T30 and RB types were related to asymptomatic samples. Based on phylogenetic analysis of the amino acid sequences of p20, p23 and p25, 19 representative CTV samples were classified into seven recently established CTV groups and a potentially novel one. A high level of genetic diversity, as well as potential recombination, was revealed among different CTV isolates. Five pure SP severe and two pure mild strains were identified by genotype composition analysis. Taken together, the results update the genetic diversity of CTV in Hunan with the detection of one possible novel strain, and this information might be applicable for the selection of appropriate mild CTV strains for controlling citrus SP disease through cross-protection.



Similar content being viewed by others
References
Albiach-Martí MR, Mawassi M, Gowda S, Satyanarayana T, Hilf ME, Shanker S, Almira EC, Vives MC, López C, Guerri J (2000) Sequences of Citrus tristeza virus separated in time and space are essentially identical. J Virol 74:6856–6865
Albiach-Marti MR, Robertson C, Gowda S, Tatineni S, Belliure B, Garnsey SM, Folimonova SY, Moreno P, Dawson WO (2010) The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3’-terminal region of the viral genome. Mol Plant Pathol 11:55–67
Broadbent P, Bevington K, Coote B (1991) Control of stem pitting of grapefruit in Australia by mild strain protection. In: Proceedings of the 11th conference on IOCV, pp 64–70
Costa AT, Nunes WMdC, Zanutto CA, Müller GW (2010) Stability of Citrus tristeza virus protective isolates in field conditions. Pesqui Agropecu Bras 45:693–700
Dawson T, Mooney P (2000) Evidence for trifoliate resistance breaking isolates of Citrus tristeza virus in New Zealand. Proceedings of the 14th conference of the IOCV. IOCV, Riverside, pp 69–76
Folimonova SY, Robertson CJ, Shilts T, Folimonov AS, Hilf ME, Garnsey SM, Dawson WO (2010) Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J Virol 84:1314–1325
Gillings M, Broadbent P, Indsto J, Lee R (1993) Characterisation of isolates and strains of citrus tristeza closterovirus using restriction analysis of the coat protein gene amplified by the polymerase chain reaction. J Virol Methods 44:305–317
Harper SJ, Dawson TE, Pearson MN (2010) Isolates of citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol 155:471–480
Harper SJ (2013) citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4
Hilf M, Garnsey S (2000) Characterization and classification of citrus tristeza virus isolates by amplification of multiple molecular markers. Proceedings of the 14th conference of international organization of citrus virologists. IOCV Riverside, CA, pp 18–27
Hilf ME, Mavrodieva VA, Garnsey SM (2005) Genetic marker analysis of a global collection of isolates of citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 95:909–917
Jiang B, Hong N, Wang GP, Hu J, Zhang JK, Wang CX, Liu Y, Fan XD (2008) Characterization of citrus tristeza virus strains from southern China based on analysis of restriction patterns and sequences of their coat protein genes. Virus Genes 37:185–192
Kong P, Rubio L, Polek M, Falk BW (2000) Population structure and genetic diversity within California citrus tristeza virus (CTV) isolates. Virus Genes 21:139–145
Licciardello G, Raspagliesi D, Bar-Joseph M, Catara AF (2012) Characterization of isolates of citrus tristeza virus by sequential analyses of enzyme immunoassays and capillary electrophoresis-single-strand conformation polymorphisms. J Virol Methods 181:139–147
Licciardello G, Xiao C, Russo M, Dai SM, Daden M, Deng ZN, Catara AF (2015) Genetic structure of citrus tristeza virus in Hunan province (PR China). Acta Hortic 1065:781–790
Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci USA 101:15742–15747
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Maydt J, Lengauer T (2006) Recco: recombination analysis using cost optimization. Bioinformatics 22:1064–1071
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS (2010) Genetic diversity and evidence for recent modular recombination in Hawaiian citrus tristeza virus. Virus Genes 40:111–118
Roy A, Brlansky R (2004) Genotype classification and molecular evidence for the presence of mixed infections in Indian citrus tristeza virus isolates. Arch Virol 149:1911–1929
Roy A, Brlansky R (2009) Population dynamics of a Florida citrus tristeza virus isolate and aphid-transmitted subisolates: identification of three genotypic groups and recombinants after aphid transmission. Phytopathology 99:1297–1306
Roy A, Brlansky R (2010) Genome analysis of an orange stem pitting citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Rubio L, MaA Ayllón, Kong P, Fernández A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Sambade A, Lopez C, Rubio L, Flores R, Guerri J, Moreno P (2003) Polymorphism of a specific region in gene p23 of citrus tristeza virus allows discrimination between mild and severe isolates. Arch Virol 148:2325–2340
Sambade A, Ambrós S, López C, Ruiz-Ruiz S, de Mendoza AH, Flores R, Guerri J, Moreno P (2007) Preferential accumulation of severe variants of citrus tristeza virus in plants co-inoculated with mild and severe variants. Arch Virol 152:1115–1126
Scott KA, Hlela Q, Zablocki O, Read D, Van-Vuuren S, Pietersen G (2013) Genotype composition of populations of grapefruit-cross-protecting citrus tristeza virus strain GFMS12 in different host plants and aphid-transmitted sub-isolates. Arch Virol 158:27–37
Soler N, Plomer M, Fagoaga C, Moreno P, Navarro L, Flores R, Pena L (2012) Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol J 10:597–608
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Weng Z, Barthelson R, Gowda S, Hilf ME, Dawson WO, Galbraith DW, Xiong Z (2007) Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. Plos One 2:e917
Wu GW, Pan S, Wang GP, Tang M, Liu Y, Yang F, Hong N (2013) The genotypes of citrus tristeza virus isolates from China revealed by sequence analysis of multiple molecular markers. Arch Virol 158:231–235
Wu GW, Tang M, Wang GP, Jin FY, Yang ZK, Cheng LJ, Hong N (2015) Genetic diversity and evolution of two capsid protein genes of citrus tristeza virus isolates from China. Arch Virol 160:787–794
Zhou C, Hailstones D, Broadbent P, Connor R, Bowyer J (2002) Studies on mild strain cross protection (MSCP) against stem-pitting tristeza virus. In: Proceedings, pp 151–157
Zhou Y, Zhou CY, Song Z, Liu KH, Yang FY (2007) Characterization of citrus tristeza virus isolates by indicators and molecular biology methods. Agr Sci China 6:573–579
Acknowledgments
The authors gratefully acknowledge Dr. Shunyuan Xiao and Dr. Yunliu Zeng for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
Financial support for this study was provided by the International Science & Technology Cooperation Project of Hunan Province (2014WK2011) and the Special Fund for Agro-Scientific Research in the Public Interest (201203076-06).
Conflict of interest
No conflict of interest.
Ethical approval
This article does not contain any studies of animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, C., Yao, RX., Li, F. et al. Population structure and diversity of citrus tristeza virus (CTV) isolates in Hunan province, China. Arch Virol 162, 409–423 (2017). https://doi.org/10.1007/s00705-016-3089-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3089-z