Abstract
Using monoclonal antibodies raised against a Faba bean necrotic yellows virus (FBNYV) isolate from Egypt and a Faba bean necrotic stunt virus (FBNSV) isolate from Ethiopia, a striking serological variability among nanovirus isolates from faba bean in Morocco was revealed. To obtain a better understanding of this nanovirus variability in Morocco, the entire genomes of two serologically contrasting isolates referred to as Mor5 and Mor23 were sequenced. The eight circular ssDNA components, each identified from Mor5- and Mor23-infected tissues and thought to form the complete nanovirus genome, ranged in size from 952 to 1,005 nt for Mor5 and from 980 to 1,004 nt for Mor23 and were structurally similar to previously described nanovirus DNAs. However, Mor5 and Mor23 differed from each other in overall nucleotide and amino acid sequences by 25 and 26%, respectively. Mor23 was most closely related to typical FBNYV isolates described earlier from Egypt and Syria, with which it shared a mean amino acid sequence identity of about 94%. On the other hand, Mor5 most closely resembled a FBNSV isolate from Ethiopia, with which it shared a mean amino acid sequence identity of approximately 89%. The serological and genetic differences observed for Mor5 and Mor23 were comparable to those observed earlier for FBNYV, FBNSV, and Milk vetch dwarf virus. Following the guidelines on nanovirus species demarcation, this suggests that Mor23 and Mor5 represent isolates of FBNYV and FBNSV, respectively. This is the first report not only on the presence of FBNSV in a country other than Ethiopia but also on the occurrence and complete genome sequences of members of two nanovirus species in the same country, thus providing evidence for faba bean crops being infected by members of two distinct nanovirus species in a restricted geographic area.
Similar content being viewed by others
Introduction
There are two families of single-stranded (ss) DNA viruses that infect plants, namely the Geminiviridae and Nanoviridae. The latter family comprises the genera Babuvirus and Nanovirus [46, 47]. Viruses of the genus Babuvirus infect monocotyledonous plants and belong to the assigned species Banana bunchy top virus (BBTV) [41] as well as to the proposed species [http://www.ictvonline.org] abaca bunchy top virus (ABTV) [40] and cardamom bushy dwarf virus (CBDV) [34]. Faba bean necrotic yellows virus (FBNYV), Milk vetch dwarf virus (MDV) and Subterranean clover stunt virus (SCSV) as well as the proposed species Faba bean necrotic stunt virus (FBNSV [17]) currently form the genus Nanovirus [46, 47]. These viruses infect dicots [9, 14, 47] and cause severe yield losses in a wide range of legume crops in Australia, Spain, and many countries of Africa and Asia [1, 9, 29, 31, 35, 47]. In contrast to geminiviruses, nano- and babuviruses are vectored by aphids. However, in common with geminiviruses, they are transmitted in a persistent, circulative and non-propagative manner [15, 47].
Members of the Nanoviridae have a multipartite ssDNA genome. Each of the circular ssDNA components appears to be individually encapsidated in a small isometric particle measuring only 18 nm in diameter. All DNAs seem to be structurally similar in being positive sense, transcribed in one direction, predominantly monocistronic, and containing a conserved stem-loop structure and other conserved domains in the noncoding region (NCR) [46]. Although up to 12 distinct DNA components have been identified from virion preparations from members of different nanovirus species, there is increasing evidence that the babuvirus genome consists of six distinct ssDNAs [8, 22, 23, 40], while the nanovirus genome comprises eight species of circular ssDNA [17, 42, 46, 47]. Babu- and nanoviruses share a set of five homologous DNA components, namely DNA-R, -S, -C, -M and -N, which code for master Rep (M-Rep), structural (capsid), cell-cycle link, movement and nuclear shuttle proteins, respectively [47]. Three other DNAs (DNA-U1, -U2 and -U4), encoding proteins whose functions are still unknown, have been identified from the nanoviruses FBNYV, MDV and FBNSV [17, 47], and one further DNA (DNA-U3) from the babuviruses BBTV [23] and ABTV [40]. In addition to these bona fide integral genome components, additional Rep-encoding DNAs have been found associated with several nano- and babuvirus isolates [20, 47], which encode distinct Rep proteins that, in contrast to the M-Rep, can only initiate the replication of their cognate DNA [18, 19, 44].
The production of 19 monoclonal antibodies (MAbs) raised against a typical FBNYV isolate from Egypt (FBNYV-Eg) not only contributed to more sensitive detection of FBNYV in plants and aphids but also permitted the identification of at least six distinct epitopes on particles of FBNYV-like nanovirus isolates [13, 15]. The observation that polyclonal antibodies to FBNYV-Eg gave weak and strong reaction with SCSV and MDV, respectively [27], and that 16 of the 19 MAbs to FBNYV-Eg cross-reacted with MDV and only one of them with SCSV [13], suggested that the serological relationship of FBNYV to MDV is close and that to SCSV is only distant. On the other hand, this also indicated that the majority of the MAbs to FBNYV-Eg are unable to discriminate FBNYV not only from MDV but presumably also from other yet unknown nanovirus species that are closely related to FBNYV. Therefore, we cannot rule out the possibility that the frequent use of these non-discriminating MAbs may have led to the erroneous serological identification of FBNYV in several Asian and African countries. The observations that several of the 19 MAbs raised against FBNYV-Eg failed to react with nanovirus isolates in faba bean samples from Ambo, Ethiopia [13], and Holetta, Ethiopia [17, 25], prompted us to sequence the genomic DNAs of the latter isolate [17, 25]. Since the DNA sequences of this isolate differed from those of FBNYV to the same extent (by 25–27%) as FBNYV differs from MDV, it has been proposed to represent a distinct nanovirus species referred to as FBNSV [17]. Furthermore, MAbs that react specifically with FBNSV but not with different FBNYV isolates were also produced for specific detection of FBNSV [17].
Morocco is one of the major faba-bean-growing countries in North Africa, where several viruses including the luteovirids pea enation mosaic virus, bean leaf roll virus and viruses belonging to the beet western yellows virus subgroup (e.g., turnip yellows virus) are among the important production constraints [10–12]. In addition, there is unconfirmed serological evidence for the occurrence of FBNYV-like (nanovirus) isolates in faba bean crops in Morocco [13, 33]. However, information on the relative importance of nanoviruses for faba bean production in Morocco is very limited [33]. Apart from the observation that nine faba bean samples from the Fez area and one faba bean sample from Meknes failed to react respectively with one and two of the 19 Mabs to FBNYV-Eg [13], none of the nanovirus isolates from Morocco has been adequately characterized.
The aforementioned epitope profiles observed for a few incidentally collected samples from Morocco in 1994 [13] indicated that the nanovirus isolates in this country differ to some extent from typical FBNYV isolates. This prompted us to use the available panel of MAbs for a serological analysis of several faba bean samples collected in Morocco in 2001. Since two of the samples gave very contrasting epitope profiles, they were selected for nucleotide sequencing. Sequence analysis of the eight ssDNAs that make up the genome of the two serologically distinct nanovirus isolates from Morocco provided strong evidence for the occurrence of two nanovirus species in this country, namely a largely typical FBNYV isolate and an FBNSV isolate distinctly different from the type isolate of FBNSV from Ethiopia.
Materials and methods
Virus isolates and serological analysis
Ten leaf samples collected from faba bean plants showing yellowing, stunting and necrosis symptoms in farmers’ fields in different areas of Morocco in 2001 were analyzed serologically by triple antibody sandwich enzyme-linked immunosorbent assay (TAS-ELISA) as described previously [13]. The geographic origin of the samples is given in Table 1. While polyclonal IgG to FBNYV [27] was used for antigen trapping, the following MAbs were used individually as detecting antibodies in TAS-ELISA:
-
1.
A mixture of three broad-spectrum MAbs (1-1F2, 2-1A1, 3-4F2) to FBNYV-like nanoviruses [13];
-
2.
MAbs 1-3D8, 2-5H9, 2-3E12-D5, and 3-4A5 raised against an FBNYV isolate from Egypt (FBNYV-Eg) but discriminating some FBNYV-like viruses [13], and
-
3.
MAbs 8-2G10, 8-4F9, 8-6F8-A5, and 8-3G11 raised against an Ethiopian isolate of FBNSV [17].
For luteovirid detection in TAS-ELISA, IgG to turnip yellows virus was used as trapping antibody for coating of plates in combination with the broad-spectrum luteovirid-specific MAb B-2-5G4 detecting antibody [24].
FBNYV-Eg was available in frozen faba bean leaves, whereas the type isolate of FBNSV [ET:Hol:97] [17] was maintained in Vicia faba seedlings of cultivar (cv) Condor and/or Scirocco by vector transmission at intervals of 3–4 weeks as described [17].
Oligonucleotide primers used and immunocaptue (IC)-PCR
For amplification of the Mor5 and Mor23 genomes, primer pair P3/P20 [43] was used to amplify a part of DNA-R selectively. Primer pairs P23 (5′-CACGAATCACAGATCCTGAT-3′)/P44 (5′-AAGCGAAHCTGACGGAAGA-3′) and P75 (5′-TAGTATTACCCCGTCCC-3′)/P76 (5′-GTAATACTAAGCCCCGTC-3′) were used to randomly amplify a part of DNA-S, -C, -M, -N, -U1, -U2, and -U4 (non-rep DNAs). Primer pairs P23/P44 and P75/P76 target conserved domains in the non-coding region of nanovirus DNAs other than DNA-R (Katul and Vetten, unpublished). In addition, several primers specific for one or several DNA components of each of the isolates were designed and used to amplify the respective DNA components randomly or selectively or to obtain the complete sequence of an already identified component.
Amplification of viral sequences was done in a 0.5-ml Eppendorf tube using an immunocapture PCR procedure described previously [16]. The PCR mixture consisted of 5 μl 10× PCR buffer, 2 mM MgCl2, 0.6 mM dNTPs, 4 μM of each primer and 1 μl Taq polymerase (Gibco, 5 U/μl). PCR was performed in a thermocycler (MJ Research, Massachusetts, USA) with a temperature profile of 5 min at 95°C for initial denaturation followed by 30 cycles of 1 min at 95°C for denaturation, 1 min at 40–65°C for annealing, depending on the primer composition, and 1 min at 72°C for extension, and followed by a final extension of 10 min at 72°C.
Cloning and analysis of restriction fragment length polymorphisms
PCR-generated DNAs were subjected to electrophoresis on a 1% agarose gel and purified using a Nucleospin® Extract kit (Macherey-Nagel, Düren, Germany). Purified DNAs were ligated into pGEM-T® vector (Promega) and used to transform competent E. coli cells (DH5α) as described [38]. Single bacterial colonies were examined for the presence of recombinant plasmids by PCR using standard T7 and SP6 vector primers. Recombinant plasmids from randomly selected positive colonies were purified (Nucleospin® plasmid kit, Macherey-Nagel, Düren, Germany), and partial nucleotide sequence information was obtained for some of the eight nanovirus DNA components. Alternatively, restriction fragment length polymorphism (RFLP) analysis was done on the SP6/T7-generated PCR products from the recombinant plasmids in an attempt to screen inserts with distinct sequences. RFLP was done with restriction enzymes BamHI, DraI, EcoRI, EcoRV, HaeIII, HindIII, HpaI, NcoI, PvuII, StyI, XbaI, XhoI, and Tth111I (Fermentas) according to the manufacturer’s instructions. These enzymes were chosen because they were expected to discriminate between each of the seven non-rep DNAs based on known sequence information from the Syrian and Egyptian isolates of FBNYV and the Ethiopian isolates of FBNSV and MDV. The resulting fragments were resolved on a 2% agarose gel to identify different restriction patterns. Recombinant plasmids containing distinct restriction recognition sites were purified and sequenced. Once a part of a given non-rep DNA had been sequenced, primers were designed based on sequence information already gained for the (specific) coding sequence of this DNA component. This permitted filling of the gaps by further PCR experiments, followed by cloning and sequencing.
Sequencing and sequence analysis
Each nucleotide of the two genomes was determined from both strands of at least two independent recombinant plasmids (i.e., from at least four independent reactions), and sequencing was performed by the custom sequencing service at Eurofins/MWG Biotech (Ebersberg, Germany). Sequence assembly and multiple alignment of nucleotide and amino acid sequences were done using the DNAMAN software package (Lynnon Biosoft, Canada). The sequences were compared with available sequences in the public genome database using BLAST analysis [4]. Phylogenetic analysis was done by multiple alignment of the sequences to homologous components of previously sequenced isolates obtained from the database using the ClustalX program [45]. The results of the multiple alignment of sequences using neighbor-joining algorithms were visualized using the default parameters of the DNAMAN software package and/or the Treeview program [36]. For sequence comparison and/or phylogenetic analysis, nanovirus sequences were retrieved from the NCBI database. For the purpose of this paper (Table 2), numbering of nucleotides starts with the third A (at the “origin of replication”) in the loop region of the highly conserved stem-loop sequence (Fig. 1).
Alignment of the replication origin sequences of the eight DNAs of the isolates Mor5 and Mor23 illustrating the extent of sequence conservation in the stem-loop common region (CR-SL). Inverted repeat sequences (open horizontal arrows) potentially forming a stem-loop structure are in bold. A vertical arrow indicates the ‘origin of replication’, which is the position of cleavage by the master Rep protein [44]. Conserved iteron-like sequences (a) and their respective inversions (a′) possibly acting as recognition or binding sites for the master Rep protein are boxed and indicated by horizontal solid arrows. Dots indicate gaps included to maximise alignments. The AatII site (GACGTC) conserved in many nanoviruses [17] but absent from the Mor23 sequences is shown in bold italics in the Mor5 sequences
Results
Epitope profiles of viruses in the Moroccan samples
Eight of the ten faba bean samples from Morocco reacted with the broad-spectrum luteovirid-specific MAb B-2-5G4 (data not shown). This demonstrated the occurrence of unidentified luteo- and/or poleroviruses in these samples, some of which have recently been identified as isolates of Turnip yellows virus [2] and Chickpea chlorotic stunt virus [3]. In addition, seven of the ten Moroccan samples reacted with the mixture of nanovirus-specific MAbs, indicating the occurrence of FBNYV-like nanoviruses in Morocco. The reaction patterns with these nanovirus-positive samples of the differentiating MAbs raised against FBNYV and FBNSV are shown in Table 1. Six of the seven nanovirus-positive samples gave strong reactions with two of the four MAbs raised against FBNYV-Eg and negative to intermediate reactions with the MAbs to FBNSV-Eth. This suggested that they are FBNYV isolates, although they differed from FBNYV-Eg in failing to react with MAb 2-3E12-D5, in not or weakly reacting with MAb 2-5H9, and in producing weak to intermediate reactions with two of the four MAbs to FBNSV-Eth. Only one sample (Mor5) reacted strongly with all four MAbs to FBNSV-Eth but failed to react with three of the four MAbs raised against FBNYV-Eg. The overall reaction pattern of Mor5 was almost indistinguishable from that of FBNSV-Eth, indicating a close serological relationship.
Detection of eight genomic DNAs from both Mor5 and Mor23
Since the epitope profiles obtained for individual samples indicated a considerable serological variation in the nanovirus(es) infecting faba bean in Morocco, Mor5 and Mor23, two isolates with contrasting epitope profiles were selected for nucleotide sequence analysis. Eight distinct circular ssDNAs were identified from both Mor5 and Mor23 and completely sequenced. The individual DNAs ranged in size from 952 to 1,005 nt for Mor5 and from 980 to 1,004 nt for Mor23 (Table 2), amounting to a genome size of 7,886 and 7,936 nt, respectively. The sizes of the individual DNAs, the properties of the potentially encoded protein, and the positions of important sequence elements are presented in Table 2. RFLP and sequence analysis (data not shown) provided no indication of the presence of DNA components other than the eight DNAs considered integral parts of the nanovirus genome [17, 46, 47].
Analysis of the noncoding regions of the Mor5 and Mor23 DNAs
The noncoding regions (NCRs) of all eight DNAs of each isolate shared a highly conserved inverted repeat sequence predicted to form a stem-loop structure that is characteristic of nanovirus DNAs [7, 26, 28, 39, 43]. An alignment of these conserved stem-loop and flanking iteron-like sequences of the Mor5 and Mor23 DNAs is shown in Fig. 1. The stem-loop sequences [nt −18 (or −20) to +13 (or +11)] of Mor5 are identical to those of FBNSV-Eth (not shown) and Mor23. Further, these three virus isolates are also indistinguishable in the component (DNA-C and -U1)-specific differences (A/G and C/T at nt positions −15 and +8, respectively) in their stem-loop sequences. The nonanucleotide sequence TAGTATT↓AC, which encompasses the origin of replication (Timchenko et al. 1999), is perfectly conserved in all the DNAs of both isolates. Moreover, the domains flanking the stem-loop region are largely conserved in all the DNAs. However, it is noteworthy that there are some specific differences between Mor5 and Mor23 in the sequences 5′ of the stem loop; for instance, none of the Mor23 DNAs has the AatII site (GACGTC), which is not only present in all the DNAs of Mor5 and FBNSV-Eth but also conserved and unique in all bona fide genomic DNAs of FBNYV, MDV and SCSV, except for two integral DNAs of SCSV (Timchenko et al. 2000; Grigoras et al. 2009). For each DNA, the position of the potential promoter (TATA box) sequence TATATAA preceding the ORFs is indicated in Table 2. Potential polyadenylation signal sequences (AATAAA), which are required for transcription termination [37], are found 3′ of the major ORF in all DNAs.
In addition to the conserved stem-loop sequence and adjacent regions referred to as common region stem-loop (CR-SL), the NCR of the seven non-rep DNA components of each isolate contained further conserved domains. This became evident from phylogenetic trees illustrating the relationship among the NCRs of the seven non-rep DNAs of Mor5 (Fig. 2a) and Mor23 (Fig. 2b). For both isolates, the NCRs of DNA-U1 and -C were most similar (96% identity), followed by those of DNA-M and -U2 (93% identity). However, these two pairs appeared to be most divergent from each other. The DNA-S and -U4 NCRs branched off from the node joining the DNA-M and -U2 NCRs, whereas the NCR of DNA-N formed a separate branch showing the least relationship to all the others. In general, the trees of the Mor5 and Mor23 NCRs had a similar topology with the exception of slight differences in the branching pattern of the DNA-S and -U4 NCRs. Since DNA-R has a large ORF and its NCR virtually contains the CR-SL only, it was excluded from this NCR comparison.
Sequence comparison of the Mor5 DNAs with the homologues of FBNSV-Eth, its closest relative, revealed that there are some variations arising from possible deletion or insertion events in the NCR. Together with all other FBNYV isolates sequenced (Eg and Sy, including Mor23), Mor5 has a stretch of 55 nucleotides in the closely related NCRs of DNA-M (nt 799–854) and -U2 (nt 797–852). Moreover, the NCR of the Mor5 DNA-N has a 21-nt insertion (nt 70–90), which is absent from the DNA-N of all the other sequenced isolates.
Analysis of the proteins potentially encoded by the Mor5 and Mor23 DNAs
Each of the eight DNAs of both Mor5 and Mor23 contains one major ORF potentially coding for a protein that is comparable in size and sequence not only to its homologue of the other isolate but also to that of all other nanovirus isolates studied so far. The ATG start codon of all of the ORFs is in a favorable context (Table 2), which at least partially agrees with the optimal translation context of plant mRNAs [30]. Key amino acid motifs such as the NTP-binding motifs (GPQ/NGGEGKT) and active site tyrosine (YxxK) of the Rep proteins [44] and LxCxE motifs of the Clink protein encoded by DNA-C [5] are conserved in their respective positions in the corresponding proteins from both isolates (data not shown).
Pairwise comparisons of the total nucleotide and encoded proteins of the two isolates were carried out with the available sequences of other isolates assigned to definitive and proposed members of the genus Nanovirus (Table 3). In agreement with the serological data, the coat protein amino acid sequences of Mor5 and Mor23 were more closely related to those of FBNSV-Eth (identity of 99%) and FBNYV-Eg (and -Sy) (97–98%), respectively. Similarly, the total nucleotide sequence DNA-S of Mor5 is most similar (88%) to that of FBNSV-Eth, while that of Mor23 has identities of 91–92% with FBNYV-Eg and -Sy. Comparisons of the other seven DNAs also showed similar relationships at both the nucleotide and amino acid level, although the degree of divergence varied considerably between the individual DNAs and their encoded proteins. Of all the nanovirus proteins, the master Rep protein encoded by DNA-R is the most conserved gene product, with identities ranging from 93 to 97% for FBNYV, FBNSV and MDV isolates and of only ~83% for SCSV. In contrast, DNA-U4 appears to encode the least conserved nanovirus protein, with identities as low as 50% between isolates of distinct nanovirus species (Table 3). All of the proteins encoded by Mor5 and Mor23 shared striking levels of sequence identity with the corresponding proteins of other nanovirus isolates (Table 3). Since, together with other nanovirus isolates, the two Moroccan isolates shared significant levels of amino acid sequence similarities with babuviruses only in DNA-R (~55%) and -N (~45%), but not in DNA-S, -M, and -C (<20%), no comparisons with babuvirus sequences are shown in Table 3.
Sequence comparison also revealed that there are some variations stemming from possible deletion or insertion events in the coding regions. Unlike FBNSV-Eth, the DNA-M ORF of Mor5 and FBNYV isolates had an 18-nt deletion at its 5′-terminus. Due to a premature stop codon, the DNA-U1 ORF of Mor5 also potentially codes for a protein that lacks 13 amino acids at its C-terminus when compared to other U1 proteins, including that of FBNSV-Eth, the closest relative of Mor5. The DNA-U2 of both Mor5 and FBNSV-Eth codes for a protein that is three amino acids shorter than the U2 protein of Mor23 or FBNYV isolates (Table 2).
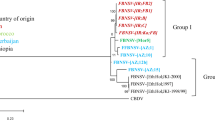
Phylogenetic analysis of all eight deduced protein amino acid sequences of Mor5 and Mor23 together with those of the other nanovirus isolates clearly demonstrated that Mor5 and Mor23 cluster with FBNSV-Eth and FBNYV-Eg (and -Sy), respectively (Fig. 3). This grouping was similar for the different proteins, although the relative position of individual isolates within each cluster varied for the different proteins, indicating different levels of conservation.
Phylogenetic trees illustrating the relationship between the eight DNAs of Mor5 (GQ274031-8) and Mor23 (GQ274023-30) and the corresponding DNAs of the Ethiopian isolate of Faba bean necrotic stunt virus [FBNSV-(ET)], a tentative nanovirus species [17], and virus isolates [from Australia (AU), Egypt (EG), Syria (SY), Iran (IR), and Japan (JP)] assigned to Faba bean necrotic yellows virus (FBNYV), Milk vetch dwarf virus (MDV), Subterranean clover stunt virus (SCSV), assigned species of the genus Nanovirus. Horizontal branch length are scaled (see bar) according to the number of base substitutions per site. Bootstrap values (1,000 replicates) higher than 70% are shown at nodes
Discussion
Our use of nanovirus broad-spectrum and specific MAbs for the serological analysis of ten faba bean samples from Morocco permitted not only the detection but also the differentiation of two nanoviruses in these samples. Based on the reaction patterns of the MAbs with the seven nanovirus-positive samples, six of them appeared to contain isolates of FBNYV, whereas only one sample (Mor5) was infected with FBNSV. The reaction pattern of Mor5 with the four MAbs raised against FBNYV-Eg was similar to that of a faba bean sample (M46) from the Meknes area in Morocco, which had also failed to react with MAbs 1-3D8 and 3-4A5 and reacted only weakly with MAbs 2-3E12-D5 and 2-5H9 [13], suggesting that an FBNSV isolate similar to Mor5 occurred in Morocco already in 1994. In contrast to Mor23, which was authenticated as an FBNYV isolate by genome sequencing, three (Mor4, Mor19, and Mor 23-2) of the FBNYV-suspected samples gave intermediate reactions with two of the FBNSV-specific MAbs. This may be an indication of the presence of FBNYV isolates that are serologically even more distinct from typical FBNYV isolates from Syria and Egypt than Mor23. The genetic distinctness of Mor4, Mor19, and Mor 23-2 remains to be shown by genome sequencing.
In a previous study of the FBNSV-Eth genome, the primer pair P3/P20 and the two primer pairs P23/P44 and P75/P76 had been used successfully for amplification of DNA-R and the seven non-rep DNAs, respectively [25]. RFLP analyses of more than 100 SP6/T7-generated PCR products from the recombinant plasmids obtained from Mor5 and Mor23 DNA with the primer pairs P23/P44 and P75/P76 and sequencing of dozens of these plasmids indicated that this experimental approach permits the detection of the seven non-rep DNAs of a nanovirus. Although the primers had been derived from a highly conserved region that is shared by all non-rep DNAs, there was no indication of the presence of a DNA other than the already known non-rep DNAs. Moreover, no additional (satellite-like) rep DNAs were encountered, as the primers used for amplification of genome components were not designed to amplify such nanovirus DNAs. This confirms the notion that the non-coding and coding sequences of the additional rep DNAs of nanoviruses differ strikingly from those of the integral DNAs of the nanovirus genome [43, 44]. The data from our study on the Mor5 and Mor23 genomes support the proposal by Vetten et al. [46] and is consistent with the recent findings of Grigoras et al. [17] that the complete genome of members of the genus Nanovirus consists of eight distinct DNAs.
Highly conserved NCR domains shared by only some of the non-rep DNAs (e.g. DNA-M and -U2 vs. DNA-C and -U1) but not by others (Fig. 2) have also been observed previously for BBTV [8], FBNYV [26, 28], MDV [39], and SCSV [7]. Hughes [21] hypothesized that these phenomena might be explained by the occurrence of repeated recombination events between the NCRs of genome components encoding non-homologous nanovirus proteins. The author further suggested that components that have been homogenized by recent genetic exchange remain quite divergent at sequences upstream or downstream of the stem-loop regions. Therefore, it is possible that a more recent recombination has taken place in the DNA-S and -U4 NCRs, which differ from the other two NCR groups despite some partial identity to each one of them.
Phylogenetically, Mor23 clusters with the isolates FBNYV-Eg and -Sy, whereas the Mor5 and FBNSV-Eth form a second group that is clearly distinct from the former group as well as from MDV and SCSV (Fig. 3). The difference in both total nucleotide and encoded amino acid sequences between the two groups is so large that the two groups should not be considered isolates or strains of the same virus. According to the current nanovirus species demarcation criteria [46], members of two distinct nanovirus species should have an overall nucleotide sequence identity of ≤75% and/or a CP amino acid sequence identity of ≤85%. The two groups share a mean nucleotide sequence identity of slightly less than 75% and a CP amino acid sequence identity of ≤85%, with a much lower mean percentage identity (ca. 74%) for all of the proteins. This is comparable to the difference between MDV and typical FBNYV isolates (Table 3). Therefore, Mor5 is proposed to be an isolate of FBNSV, whereas Mor23 is an FBNYV isolate. It remains to be shown whether there are also consistent differences between FBNYV and FBNSV isolates in host range, symptomatology, and aphid vector specificities.
The results presented here provide the first evidence for the occurrence of both FBNYV and FBNSV in the same country. While FBNYV has been reported from Egypt and Syria [26–28, 32] and unconfirmed serological data suggest that it occurs in several African and Asian countries [1, 13, 27, 31, 33], FBNSV has been reported previously only from Ethiopia [17, 25]. Because of the serological and molecular variability observed for FBNYV-like isolates from Ethiopia and other countries, Franz et al. [13, 16] and Katul and Vetten [25] explained such observations as a geographically associated variation among FBNYV-like nanoviruses from different countries. However, the results of this study provide a more complex picture of nanovirus variability in a country, because they demonstrated that both FBNYV and FBNSV, which belong to two distinct nanovirus species, infect faba bean in a single country. A similar observation has recently been made for Ethiopia, where three distinct nanoviruses can coexist in the same geographic area (Abraham et al. unpublished).
The Moroccan FBNSV isolate Mor5 was genetically quite distinct from the Ethiopian FBNSV isolate, from which it was serologically (and in coat protein sequences almost) indistinguishable, thus providing again an indication of a possible geographically associated variation among isolates of a nanovirus species. Intensive future surveys in other countries are therefore likely to reveal the occurrence of more nanovirus species and further nanovirus strains. Most of the information available so far on nano- and babuvirus variability is for BBTV, for which sequence information on the DNA-R, -N, and -S of isolates from different countries has revealed the occurrence of two geographical groups: Asian and South Pacific isolates corresponding to their geographical origin [22, 23, 48]. A more detailed study on DNA-R sequences of BBTV isolates from Vietnam [6] indicated that although all isolates fall within the Asian group, there was a marked difference between isolates from southern and northern Vietnam. Similarly, striking genetic differences ranging from 7 to 20% were also observed for the individual gene products of two ABTV isolates, one from the Philippines and the other from Malaysia [40]. Although the number of samples used in our study did not allow us to comment on the geographical distribution of FBNYV and FBNSV isolates in Morocco, our study is the first that provides genome sequence information on two nanovirus isolates infecting faba bean in a single country.
The occurrence of two distinct nanovirus species infecting faba bean in a restricted geographic area may have implications for future attempts at breeding for resistance to the viruses. In countries like Morocco, where two nanovirus species are now known to occur, it is possible that resistance targeted to only one species may not be effective against the other. More surveys are required for determining the occurrence, geographical distribution and relative importance of these two nanovirus species not only in Morocco but also in other countries of West Asia and North Africa.
References
Abraham A, Makkouk KM, Gorfu D, Lencho AG, Ali K, Tadesse N, Yusuf A, Lencho A (2000) Survey of faba bean (Vicia faba L.) virus diseases in Ethiopia. Phytopath Mediterr 39:277–282
Abraham AD, Varrelmann M, Vetten HJ (2008) Molecular evidence for the occurrence of two new luteoviruses in cool season food legumes in Northeast Africa. African J Biotechnol 7:414–420
Abraham AD, Menzel W, Varrelmann M, Vetten HJ (2009) Molecular, serological and biological variation among chickpea chlorotic stunt virus isolates from five countries of North Africa and West Asia. Arch Virol 154:791–799
Altschul SF, Madden TL, Schaffer AA, Zhang ZH, Zhang Z, Miller WW, Lippman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aronson MN, Meyer AD, Gyorgyey J, Katul L, Vetten HJ, Gronenborn B, Timchenko T (2000) Clink, a nanovirus encoded-protein binds both pRB and SKP1. J Virol 74:2967–2972
Bell KE, Dale JL, Ha CV, Vu MT, Revil PA (2002) Characterization of Rep-encoding components associated with banana bunchy top nanovirus in Vietnam. Arch Virol 147:695–707
Boevink P, Chu PWG, Keese P (1995) Sequence of subterranean clover stunt virus DNA: affinities with the geminiviruses. Virology 207:354–361
Burns TM, Harding RM, Dale JL (1995) The genome organization of banana bunchy top virus: analysis of six ssDNA components. J Gen Virol 76:1471–1482
Chu PWG, Vetten HJ (2003) Subterranean clover stunt virus. AAB descriptions of plant viruses no. 396. http://dpvweb.net/dpv/showadpv.php?dpvno=396
Fortass MF, Bos L (1991) Survey of faba bean (Vicia faba L) for viruses in Morocco. Neth J Pl Path 97:369–380
Fortass MF, van der Wilk F, Goldbach RW, Bos L, van den Heuvel JFJM (1996) Diversity of viruses infecting faba bean in Morocco and their detection by the polymerase reaction. Agronomie 16:61–68
Fortass MF, van der Wilk F, van den Heuvel JFJM, Goldbach RW (1997) Molecular evidence for the occurrence of beet western yellows virus on chickpea in Morocco. Eur J Plant Path 103:481–484
Franz A, Makkouk KM, Katul L, Vetten HJ (1996) Monoclonal antibodies for the detection and differentiation of faba bean necrotic yellows virus isolates. Ann Appl Biol 128:255–2268
Franz A, Makkouk KM, Vetten HJ (1997) Host range of faba bean necrotic yellows virus and potential yield loss in infected faba bean. Phytopath Med 36:94–103
Franz A, Makkouk KM, Vetten HJ (1998) Acquisition, retention and transmission of faba bean necrotic yellows virus by two of its aphid vectors, Aphis craccivora Koch and Acyrthosiphon pisum Harris. J Phytopath 146:347–355
Franz A, van der Wilk F, Verbeek M, Dullemans AM, van den Heuvel JFJM (1999) Faba bean necrotic yellows virus (genus Nanovirus) requires a helper factor for its aphid transmission. Virology 262:219–219
Grigoras I, Timchenko T, Katul L, Grande-Pérez A, Vetten HJ, Gronenborn B (2009) Reconstitution of authentic nanovirus from multiple cloned DNAs. J Virol 83:10778–10787
Horser CL, Harding RM, Dale JL (2001) Banana bunchy top nanovirus DNA-1 encodes the ‘master’ replication initiation protein. J Gen Virol 82:459–464
Horser CL, Karan M, Harding RM, Dale JL (2001) Additional Rep-encoding DNAs associated with banana bunchy top virus. Arch Virol 146:71–86
Hu JM, Fu HC, Lin CH, Su HJ, Yeh HH (2007) Reassortment and concerted evolution in Banana bunchy top virus genomes. J Virol 81:1746–1761
Hughes AL (2004) Birth-and-death evolution of protein coding regions and concerted evolution of non-coding regions in the multi-component genomes of nanoviruses. Mol Phylogenet Evol 30:287–294
Karan M, Harding MR, Dale JL (1994) Evidence for two groups of banana bunchy top virus isolates. J Gen Virol 75:3541–3546
Karan M, Harding RM, Dale JL (1997) Association of banana bunchy top virus DNA components 2 to 6 with bunchy top disease. Molecular Plant Pathology On-line (http://www.bspp.org.uk/mppol/1997/0624karan/index.htm)
Katul L (1992) Serological and molecular characterization of bean leaf roll virus (BLRV) and faba bean necrotic yellows virus (FBNYV) (in German). Ph.D. dissertation, University of Göttingen, Germany, 115 pp
Katul L, Vetten HJ (1999) Sequence analysis of an Ethiopian isolate of Faba bean necrotic yellows virus: evidence for virus variability or the presence of another nanovirus. XIth International congress of virology, Sydney, Australia, Aug 9–13, 1999, Abstract no. VP66.21, pp 240–241
Katul L, Maiss E, Morozov SY, Vetten HJ (1997) Analysis of six DNA components of the faba bean necrotic yellows virus genome and their structural affinity to related plant virus genomes. Virology 233:247–259
Katul L, Vetten HJ, Maiss E, Makkouk KM, Lesemann D-E, Casper R (1993) Characterization and serology of virus-like particles associated with faba bean necrotic yellows. Ann Appl Biol 123:629–647
Katul L, Timchenko T, Gronenborn B, Vetten HJ (1998) Ten distinct ssDNA components, four of which encode putative replication-associated proteins, are associated with the faba bean necrotic yellows virus genome. J Gen Virol 79:3101–3109
Kumari SG, Rodoni B, Vetten HJ, Loh MH, Freeman A, van Leur J, Shiying B, Xiaoming W (2009) Detection and partial characterization of Milk vetch dwarf virus isolates from faba bean (Vicia faba L.) in Yunnan Province, China. J Phytopathol (published online ahead of print on Apr 21, 2009; doi:10.1111/j.1439-0434.2009.01572.x)
Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Sheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6:43–48
Makkouk KM, Kumari SG, Hughes Jd’A, Muniyappa V, Kulkarni NK (2003) Other legumes, pp 447–476. In: Loebenstein G, Thottappilly G (eds) Virus and virus-like diseases of major crops in developing countries. Kluwer, Dordrecht, 800 pp
Makkouk KM, Rikallah L, Madkour M, El-Sherbeny M, Kumar SG, Amriti AW, Solh MB (1994) Survey of faba bean (Vicia faba L) for viruses in Egypt. Phytopath Medit 33:207–211
Makkouk KM, Kumari SG (2009) Epidemiology and integrated management of persistently transmitted aphid-borne viruses of legume and cereal crops in West Asia and North Africa. Virus Res 141:209–218
Mandal B, Mandal S, Pun KB, Varma A (2004) First report of the association of a nanovirus with foorkey disease of large cardamom in India. Plant Dis 88:428
Ortiz V, Navarro E, Castro S, Carazo G, Romero J (2006) Incidence and transmission of Faba bean necrotic yellows virus (FBNYV) in Spain. Spanish J Agric Res 4:255–260
Page GD (1996) Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Rothnie HM, Reid J, Hohn T (1994) The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′ end formation in plants. EMBO J 13:2200–2210
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sano Y, Wada M, Hashimoto Y, Matsumoto T, Kojima M (1998) Sequence of ten circular ssDNA components associated with the milk vetch dwarf virus genome. J Gen Virol 79:3111–3118
Sharman M, Thomas JE, Skabo S, Holton TA (2008) Abacá bunchy top virus, a new member of the genus Babuvirus (family Nanoviridae). Arch Virol 153:135–147
Thomas JE (2008) Banana bunchy top virus. In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of virology, vol 1, 3rd edn. Elsevier, Oxford, pp 272–279
Timchenko T, Katul L, Aronson M, Vega-Arreguin RamirezBC, Vetten HJ, Gronenborn B (2006) Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J Gen Virol 87:1735–1743
Timchenko T, Katul L, Sano Y, de Kouchkovsky F, Vetten HJ, Gronenborn B (2000) The master rep concept in nanovirus replication: identification of missing genome components and potential for natural genetic reassortment. Virology 274:189–195
Timchenko T, de Kouchkovsky F, Katul L, David C, Vetten HJ, Gronenborn B (1999) A single rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single stranded DNA virus of plants. J Virol 73:10173–10182
Thompson JD, Gibson TB, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucl Acids Res 25:4876–4882
Vetten HJ, Chu PWG, Dale JL, Harding R, Hu J, Katul L, Kojima M, Randles JW, Sano Y, Thomas JE (2005) Nanoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, pp 343–352
Vetten HJ (2008). Nanoviruses. In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of virology, vol 3, 3rd edn. Elsevier, Oxford, pp 385–391
Wanitchakorn R, Harding RM, Dale JL (2000) Sequence variability in the coat protein gene of two groups of banana bunchy top isolates. Arch Virol 145:593–602
Acknowledgments
A. D. Abraham and B. Bencharki were financially supported by the German Academic Exchange Service (DAAD). We thank Ioana Grigoras, Tania Timchenko and Bruno Gronenborn for useful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequences reported here have been assigned the GenBank acc. no. GQ274023 to GQ274030 (FBNYV-Mor23) and GQ274031 to GQ274038 (FBNSV-Mor5).
An erratum to this article can be found at http://dx.doi.org/10.1007/s00705-010-0796-8
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Abraham, A.D., Bencharki, B., Torok, V. et al. Two distinct nanovirus species infecting faba bean in Morocco. Arch Virol 155, 37–46 (2010). https://doi.org/10.1007/s00705-009-0548-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0548-9