Abstract
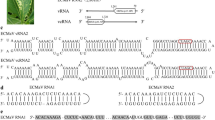
The complete sequence of four viral RNA segments of fig mosaic virus (FMV) was determined. Each of the four RNAs comprises a single open reading frame (ORF) 7,093, 2,252, 1,490 and 1,472 nucleotides in size, respectively. These ORFs encode the following proteins in the order: RNA-dependent RNA polymerase (p1 264 kDa), a putative glycoprotein (p2 73 kDa), a putative nucleocapsid protein (p3 35 kDa) and a protein with unknown function (p4 40.5 kDa). All RNA segments possess untranslated regions containing at the 5′ and 3′ termini a 13-nt complementary sequence. A conserved motif denoted premotif A was found to be present in addition to the five RdRp motifs A–F in RNA-1. In phylogenetic trees constructed with the amino acid sequences of RNA-1 and RNA-2, FMV clustered consistently with European mountain ash ringspot-associated virus (EMARaV) in a clade close to those comprising members of the genera Hantavirus, Orthobunyavirus and Tospovirus. The amino acid sequence of the putative FMV nucleocapsid protein encoded by RNA-3 shared identity with comparable sequences of EMARaV and the unclassified viruses pigeonpea sterility mosaic virus (PPSMV) and maize red stripe virus (MRSV). The nucleocapsid sequences rooted the four viruses in a clade close to the genus Tospovirus. Based on molecular, morphological and epidemiological features, FMV appears to be very closely related to PPSMV and MRSV. All these viruses are phylogenetically related to EMARaV and therefore seem to be eligible for classification in the proposed genus Emaravirus, which, in turn, may find a taxonomic allocation in the family Bunyaviridae.






Similar content being viewed by others
References
Ahn KK, Kim KS, Gergerich RC, Jensen S, Anderson EJ (1996) Comparative ltrastructure of double membrane-bound particles and inclusions associated with eriophyid mite-borne plant disease of unknown etiology: a potentially new group of plant viruses. J Submicrosc Cytol Pathol 28:345–355
Altschul SF, Stephen F, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Appiano A, Conti M, Zini N (1995) Cytopathological study of the double-membrane bodies occurring in fig plants affected by fig mosaic disease. Acta Hortic 386:585–592
Benthack W, Mielke N, Buttner C, Muhlbach HP (2005) Double-stranded RNA pattern and partial sequence data indicate plant virus infection associated with ringspot disease of European mountain ash (Sorbus aucuparia L.). Arch Virol 150:37–52
Bradfute OR, Whitmoyer RE, Nault RL (1970) Ultrastructure of plant leaf tissue infected with mite-borne viral-like particles. Proc Electron Microsc Soc Am 258:178–179
Bruenn JA (2003) A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res 31(7):1821–1829
Castellano MA, Gattoni G, Minafra A, Conti M, Martelli GP (2007) Fig mosaic in Mexico and South Africa. J Plant Pathol 89:441–443
Chomczynski P (1992) One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem 201:134–139
Condit IJ, Horne WT (1933) A mosaic of the fig in California. Phytopathology 23:887–896
Coutts RHA, Livieratos IC (2003) A rapid method for sequencing the 5′- and 3′-termini of double-stranded RNA viral templates using RLM-RACE. J Phytopathol 151:525–527
Dodds JA (1993) DsRNA in diagnosis. In: Matthews REF (ed) Diagnosis of plant virus diseases. CRC Press, Boca Raton, pp 273–294
Ebrahim-Nesbat F, Izadpanah K (1992) Virus-like particles associated with ringfleck mosaic of mountain ash and a mosaic disease of raspberry in the Bavarian forest. Eur J Forest Pathol 22:1–10
Elbeaino T, Digiaro M, Alabdullah A, De Stradis A, Minafra A, Mielke N, Castellano MA, Martelli GP (2009) A multipartite single-stranded negative-sense RNA virus is the putative agent of fig mosaic disease. J Gen Virol 90(5):1281–1288
Emanuelsson O, Nielsen H, Brunak S, Von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Emanuelsson O, Brunak S, Von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971
Flock RA, Wallace JM (1955) Transmission of fig mosaic by the eriophyid mite Aceria ficus. Phytopathology 45:52–54
Foissac X, Svanella-Dumas L, Gentit P, Dulucq MJ, Candresse T (2001) Polyvalent detection of fruit tree Tricho, Capillo and Foveavirus by nested RT-PCR using degenerated and inosine containing primers (DOP RT-PCR). Acta Hortic 550:37–43
Kulkarni NK, Lava Kumar P (2002) Transmission of Pigeon pea sterility mosaic virus by the eriophyid mite, Aceria cajani (Acari: Arthropoda). Plant Dis 86:1297–1302
Lava Kumar PL, Duncan GC, Roberts IM, Jones AT, Reddy DVR (2002) Cytopathology of Pigeonpea sterility mosaic virus in pigeonpea and Nicotiana benthamiana: similarities with those of eriophyid mite-borne agents of undefined aetiology. Ann Appl Biol 140:87–96
Lava Kumar PL, Jones AT, Reddy DVR (2003) A novel mite-transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology 93:71–81
Marck C (1988) DNA Strider: a “C” programme for the fast analysis of DNA and protein sequences on the Apple Macintosh family computers. Nucleic Acids Res 16:1829–1836
Martelli GP (2009). Fig mosaic disease and associated viruses. In: Hadidi A, Barba M, Candresse T, Jelkmann W (eds) Virus and virus-like diseases of pome and stone fruits. APS Press, St. Paul (in press)
Martelli GP, Castellano MA, Lafortezza R (1993) An ultrastructural study of fig mosaic. Phytopathol Mediterr 32:33–43
Matsuda S, Vert JP, Saigo H, Ueda N, Toh H, Akutsu T (2005) A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci 14:2804–2813
Mielke N, Muehlbach HP (2007) A novel, multipartite, negative-strand RNA virus is associated with the ringspot disease of European mountain ash (Sorbus aucuparia L.). J Gen Virol 88:1337–1346
Müller R, Poch O, Delarue M, Bishop DHL, Bouloy M (1994) Rift valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75:1345–1352
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Perlman D, Halvorson HO (1983) A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol 167:391–409
Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Proeseler G (1972) Beziehungen zwischen Virus, Vektor und Wirstpflanze am Beispiel des Feigenmosaik-Virus und Aceria ficus Cotte (Eriophyoidea). Acta Phytopathologica Academiae Scientiarium Hungaricae 7:106–179
Saldarelli P, Minafra A, Martelli GP, Walter B (1994) Detection of grapevine leafroll-associated closterovirus III by molecular hybridization. Plant Pathol 43:91–96
Skare JM, Wijkamp I, Rezende JAM, Kitajima EW, Park JW, Desvoyes B, Rush CM, Michels G, Scholthof KBG, Scholthof HB (2006) A new eriophyid mite-borne membrane-enveloped virus-like complex isolated from plants. Virology 347:343–353
Von Heijne G (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690
Von Heijne G (1983) Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem 133:17–21
Walia JJ, Salem NM, Falk BW (2009) Partial sequence and survey analysis identify a multipartite, negative-sense RNA virus associated with fig mosaic. Plant Dis 93:4–10
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Elbeaino, T., Digiaro, M. & Martelli, G.P. Complete nucleotide sequence of four RNA segments of fig mosaic virus. Arch Virol 154, 1719–1727 (2009). https://doi.org/10.1007/s00705-009-0509-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0509-3