Abstract
The immune response to CSFV and the strategies of this virus to evade and suppress the pigs’ immune system are still poorly understood. Therefore, we investigated the transcriptional response in the tonsils, median retropharyngeal lymph node (MRLN), and spleen of pigs infected with CSFV strains of similar origin with high, moderate, and low virulence. Using a porcine spleen/intestinal cDNA microarray, expression levels in RNA pools prepared from infected tissue at 3 dpi (three pigs per virus strain) were compared to levels in pools prepared from uninfected homologue tissues (nine pigs). A total of 44 genes were found to be differentially expressed. The genes were functionally clustered in six groups: innate and adaptive immune response, interferon-regulated genes, apoptosis, ubiquitin-mediated proteolysis, oxidative phosphorylation and cytoskeleton. Significant up-regulation of three IFN-γ-induced genes in the MRLNs of pigs infected with the low virulence strain was the only clear qualitative difference in gene expression observed between the strains with high, moderate and low virulence. Real-time PCR analysis of four response genes in all individual samples largely confirmed the microarray data at 3 dpi. Additional PCR analysis of infected tonsil, MRLN, and spleen samples collected at 7 and 10 dpi indicated that the strong induction of expression of the antiviral response genes chemokine CXCL10 and 2′–5′ oligoadenylate synthetase 2, and of the TNF-related apoptosis-inducing ligand (TRAIL) gene at 3 dpi, decreased to lower levels at 7 and 10 dpi. For the highly and moderately virulent strains, this decrease in antiviral and apoptotic gene expression coincided with higher levels of virus in these immune tissues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Classical swine fever virus (CSFV), a member of the genus Pestivirus, family Flaviviridae, causes a severe disease of pigs that is characterized by fever, leucopenia and hemorrhage [65, 66]. Apoptosis has been demonstrated to be the cause of lymphocyte death [58, 66], but the precise mechanism by which CSFV causes apoptosis is still unknown.
Innate immunity is a critical arm of pre-existing host defence that functions to eliminate foreign pathogens immediately after infection and subsequently directs the adaptive immune response. Interferons (IFNs) are key components of innate immunity. Through different signal-transduction pathways, IFNs can promote the expression of dozens of genes responsible for executing IFN-induced anti-viral and anti-proliferative effects (reviewed in Ref. [30]).
The best-characterized IFN-inducible components of the antiviral response are the dsRNA-dependent protein kinase R (PKR, alias eukaryotic translation initiation factor 2-alpha kinase 2), the 2′–5′ oligoadenylate synthetases (OASs), the Mx proteins and the nitric oxide system (NO). IFNs can control apoptosis by inducing a pro-apoptotic state in uninfected cells For example, IFN has been shown to influence the sensitivity of cells for apoptosis by inducing expression of both the ligand and the receptor of FAS and TRAIL [71, 73]. In addition, IFNs can control the T helper immune system by inducing a broad spectrum of chemokines and cytokines such as CXCL10, which induce the Th1 immune response [18].
Viruses employ numerous evasive strategies to impair antiviral responses. In the case of CSFV, only a few viral strategies have been identified. The CSFV Npro protein has been described as a suppressor of IRF3 function in the IFN expression pathway in infected cells [3, 7, 17, 42, 55, 56, 60] and has been shown to interact with IkBα, the NF-KB inhibitor [23]. In addition, in CSFV-infected macrophages, NO production is suppressed by up-regulation of arginase activity [75]. However, those viral mechanisms only partially explain the immune suppression caused by CSFV. Therefore, CSFV most likely uses several other strategies to impair different immune pathways, e.g., the function of antigen-presenting cells, the regulation of apoptosis, anti-viral responses and the complement system. These mechanisms have not yet been investigated for CSFV, and most of the current knowledge has been gained from in vitro experiments.
In this present study, we used micro-array and PCR analysis to investigate the transcriptional response to replication of CSFV in relevant immune organs of pigs. Pigs were infected with three different laboratory-derived variants of CSF virus strain Brescia [36], which were characterized as highly virulent, moderately virulent, and avirulent in earlier animal experiments [69]. However, because the avirulent strain did cause some fever and leucopenia, we will refer to it in this paper as low virulent. We identified viral modulations of IFN-inducible antiviral, apoptotic and chemotactic genes. To our knowledge, these data represent the first in vivo gene expression study that describes antiviral responses to CSFV.
Materials and methods
Study design
An animal experiment was carried out with four groups of nine pigs each. Three groups were inoculated with the highly, moderately or low virulent CSF strain, while the fourth group was left untreated. In each group, three piglets were euthanized at 3, 7 and 10 days post-inoculation (dpi) for the collection of tissue samples.
Animals
Thirty-six 6-week-old piglets were obtained from a conventional herd that was free of pestiviruses. Groups of four piglets of the same sex and originating from the same litter were selected from the herd. From each group of four, one piglet was randomly assigned to one of the four treatment groups.
Inoculation
On day 0, the piglets of groups 1–3 were inoculated intranasally with 2 ml virus suspension (1 ml per nostril) containing 105 TCID50 per ml of infectious virus, as determined using SK6 cells. The viruses used were, respectively, CoBrB (highly virulent: HV), C1.1.1 (low virulent: LV) and C1.1.1/CoBrB (moderately virulent: MV). Virus C1.1.1 is a cell-adapted variant of a Brescia strain with low virulence, whereas CoBrB (CoBrB 476S) is the virulent variant. Sequence analysis showed 29 nucleic acid mutations in C1.1.1, resulting in nine amino acid substitutions, compared to the sequence of CoBrB [36]. C1.1.1/CoBrB is a chimera from C1.1.1 and CoBrB, essentially containing the structural proteins from C1.1.1 and the non-structural proteins from CoBrB [69]. The control group was inoculated with 2 ml PBS instead.
Observation and scoring
From 3 days before the inoculation until they were euthanized, the animals were observed on a daily basis for clinical symptoms, and their rectal temperature was measured. A list of CSF-related clinical symptoms (lack of appetite, diarrhoea, lethargy, inability to stand up, trembling) was used and scored as either present or not present. The scores for each pig were added up to a total score per day.
Leucocyte count
EDTA blood samples were taken on days 0, 1, 2, 3, 4, 7, and 10 from all pigs that were still alive. White blood cell (leucocytes) and thrombocyte counts were performed using a Medonic® CA 620 coulter counter (Boule Medical AB, Stockholm, Sweden). Leucopenia was defined as <8 × 109 cells/l blood, and thrombocytopenia as <200 × 109 cells/l blood [12].
Post-mortem sampling
In each group, the first three piglets were euthanized on the first day on which the rectal body temperature was >40°C. At 7 and 10 dpi, another three piglets were euthanized in each group. Post-mortem tissue samples were collected from the tonsils, spleen and median retropharyngeal lymph node (MRLN). They were snap-frozen in liquid nitrogen and stored at −80°C until total RNA was isolated from these samples.
Isolation of total RNA
From 0.5 g of tissue, total RNA (DNase-free) was isolated using TRIzol® reagent (Invitrogen) as described recently [50]. The yield and purity of the RNA was calculated from measurements of the extinction at 260 and 280 nm. The integrity of all RNA samples was checked by analyzing 0.5 µg of RNA on a 1% (w/v) agarose gel. After ethidium bromide staining, the gel was scanned to calculate the 28S/18S peak ratio (volume 28S over volume 18S) for each RNA preparation. RNA with a ratio >2 was considered of adequate quality to be used for real-time PCR and microarray analysis. For all RNA preparations, a 28S/18S ratio of >2 was observed. A part of each RNA preparation was used to prepare RNA pools for microarray analysis.
Microarray analysis
For microarray analysis a homemade porcine cDNA microarray containing 2,928 probes from a jejunum EST library [50] plus 2,880 probes from a spleen EST library was used. For production of this spleen EST library, spleens were collected from the same four 12-week-old pigs from which the jejunal mucosal scrapings were obtained for the preparation of the jejunum EST library [50]. A total of 2,688 probes (ESTs) were generated from total RNA isolated from pooled spleen tissue (n = 4 pigs). In addition, 192 probes were generated from RNA isolated from in vitro ConA/LPS-stimulated spleen cells derived from the above-described pooled spleen tissue. Together with the jejunum and spleen probes, an additional 192 cDNA probes selected from Marc1 and 2 EST libraries [24], and probes coding for porcine cytokines (IFN-γ, TNF-α, GMCSF, IL-2, 4, 6, 8, and 10) and lung surfactant proteins SFTPA and SFTPD were spotted in quadruplicate on Corning UltraGAPS slides [31].
Dual-colour hybridization of slides was performed using an RNA MICROMAX TSA labelling and detection kit (Perkin-Elmer, Zoetermeer, The Netherlands) as described earlier [50]. Briefly, a biotin (BI)-labeled cDNA target was prepared from 1 µg of RNA template from an uninfected RNA pool, and a fluorescein (FL)-labeled cDNA target was prepared from 1 µg of RNA template from an infected RNA pool. Labelled cDNAs were simultaneously hybridized to a microarray slide and detected with Cy5 (biotin) and Cy3 (fluorescein), respectively. For each comparison, a second hybridization experiment was performed in which the BI and FL labels were reversed (dye-swap). The slides were scanned for Cy5 and Cy3 fluorescence in a Packard Bioscience BioChip Technologies apparatus (ScanArray Express, Perkin-Elmer). Scan images were processed with GenePix Pro 5.0 (Molecular Devices, Apeldoorn, The Netherlands), automatically gridding the spots, and measuring spot intensities. In addition, spots with irregular shape were eliminated manually. An intensity-dependent normalisation was performed (blank-specific background correction; Lowess fit function with a fraction of 0.2 on all data points) using a customized version of the statistical software package R [74]. Significantly differentially expressed spots with a ratio of >3.3 or <0.30 (M-value of <−1.73 or >1.73; M = log2 (Cy5/Cy3)) and a p-value <0.05 were selected manually from normalized data reports. A probe was considered differentially expressed when at least five out of the eight spots present on the two dye-swaps passed these selection criteria. For each probe, the mean ratio of differential expression (infected over uninfected) of replicate spots (n ≥ 5 and ≤8) is presented. Probes that were yet not annotated were sequenced as described previously [50].
Microarray comparisons
For each type of tissue, an RNA pool was prepared from total RNA, isolated from three pigs originating from the same infected group. At 3 dpi, these three piglets were selected from the HV and MV groups on the basis of early signs of fever (i.e., highest rectal temperature), and randomly from the LV group (see above). In three separate dye-swap hybridization experiments, mRNA expression levels in infected spleen, tonsil, and lymph node pools were compared to levels in RNA pools prepared from homologue tissue collected from the nine uninfected (control) pigs. Performing these tissue-specific microarrays for each virus strain (HV, MV, LV) resulted in nine data sets of probes hybridizing differentially with a fold change (ratio infected over uninfected) of <0.30 or >3.3 (p-value <0.05). Sequences of selected probes were annotated after blastn or (t)blastx analysis (when not yet annotated).
Real-time PCR for CSFV
For the detection of CFSV, PCR assays were performed using a LightCycler instrument with red dye, using the RNA Master Hybprobes kit (Roche Applied Science, Mannheim, Germany) [70]. C t values were used to express the amounts of virus in a semi-quantitative way.
Real-time PCR for gene expression
Gene-specific primers were developed from mRNA reference sequences (pig, cattle, human) found in sequence databases (NBCI, KEGG). These reference sequences were selected by homology search with EST sequences obtained from the library probes. Primers were designed within the open frame of the protein encoded by the gene of interest and also, when possible, in different exons in order to prevent amplification of genomic DNA. The primer sequences are listed in Table 1.
Randomly primed reverse transcription reactions were carried out using the Transcriptor first-strand cDNA synthesis kit (Roche Applied Science, Mannheim, Germany) with approximately 200 ng of RNA as template and 60 µM hexamer primers. The produced cDNA was stored at −20°C until use. Real-time PCR tests were carried out with a LightCycler instrument and a LightCycler FastStart DNA Master SYBR Green I kit (Roche Applied Science, Mannheim, Germany). The PCR conditions are listed in Table 1. The relative concentration of mRNA was calculated by extrapolation on a standard curve prepared from dilutions of an RT reaction prepared from a reference RNA sample. The relative concentration of 18S and beta-actin were determined once for each cDNA sample and used to normalize concentrations of all other genes tested from the same cDNA sample. Established quantities of 18S rRNA and beta-actin showed essentially no differences among any of the individual RNA samples (18S RNA average relative concentration 1.15 ± 0.30; beta-actin average relative concentration 0.54 ± 0.026).
Results
Clinical parameters after infection with CSFV strains
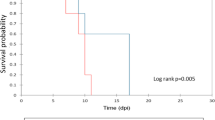
Pigs infected with the HV and MV virus strains developed fever (above 40°C) around 2–3 dpi together with a drop in WBC count and, this was then followed by acute clinical signs beginning 4 dpi (Fig. 1). Although the drop in WBC’s and fever for LV-infected pigs coincided with those of MV and HV-infected pigs, WBC concentrations and rectal temperatures reached normal levels at 7 dpi. Moreover, the moderate clinical signs observed in these pigs dramatically declined from 8 until 10 dpi. These results indicated that LV-infected pigs quickly recovered from the infection. The pigs from the control group showed normal clinical parameters during the whole animal experiment (Fig. 1).
Gene expression analysis
Messenger RNA levels in CSFV-infected immune organs at the port of entrance (tonsil and medial retropharyngeal lymph node) and in the spleen at 3 dpi were compared to levels in homologous tissues from uninfected pigs for all three virus strains (LV, MV, and HV), using the homemade cDNA microarray prepared from spleen and jejunum tissue. In these nine microarray comparisons, a total of 44 different mRNAs (genes) hybridized differentially with a ratio (fold change; infected over uninfected) of <0.30 or >3.3 (p-value <0.05). Messenger RNA levels were increased significantly for 33 genes and reduced significantly for 11 genes. In Table 2, the ratio of differential expression calculated from the two dye-swap slides is presented. Based on literature search and data mining, a tentative function was assigned for the genes identified by blast analysis. With the probes coding for IFN-γ, TNF-α, GM-CSF, IL-2, 4, 6, 8, and 10, the fluorescence intensity was never higher than the background threshold.
Data mining and functional clustering
Based on their function, genes identified by microarray analysis were clustered in six groups (Table 2): (1) genes related to apoptosis (Apopt.), (2) IFN-α/β- or γ-induced genes (IFN), (3) genes related to the adaptive and innate immune response, including antigen presentation (innate/adapt. Immune-AP), (4) genes related to the proteosome pathway, i.e., ubiquitin-mediated proteolysis (UBM), (5) components of, or proteins associated with the cytoskeleton (cytoskeleton), and (6) genes involved in oxidative phosphorylation, including oxygen transporters (Ox-P).
Up-regulated genes related to apoptosis included pro-apoptotic genes such as TRAIL, which leads to caspase-dependent cell death (reviewed in Ref. [64]) and the gene for cathepsin C, a protease responsible for the proteolytic maturation of granzyme [51], which induces apoptosis through activation of caspase and the Bid protein [8, 11]. Anti-apoptotic genes were also induced, such as MCL1 (alias BCL2-related), which blocks apoptosis mediated by ER-Ca2+ release and mitochondrial Cyt-c release [14], and is also involved in the regulation of redox- and oxidant-mediated apoptosis [32, 43]. In addition, genes related to glutathione metabolism (GLRX, GCLM), which also prevent redox- and oxidant-mediated apoptosis in the ER and mitochondria, were up-regulated [32]. Calbindin, a calcium-binding protein suppresses apoptosis by inhibiting caspase 3 activity [5]. Although PARP genes are well established as cell survival factors contributing to DNA repair, recent work has indicated that they also induce cell death via the p53 signalling pathway and via the intrinsic pathway [16, 33].
IFN-regulated genes were all up-regulated. These genes included IFN-γ-induced genes AIF-1, IFI 16, GBP-2, and MOP-5, and genes induced by IFN-α/β: IFI44, CXCL10, and OAS2. CXCL10 (alias IP10) is a pleiotropic cytokine that stimulates monocytes, natural killer and T cell migration [18]. The antiviral enzyme OAS2 catalyses the synthesis of oligoadenylate chains in response to dsRNA binding, which acts as an activator of RNase L [61]. Active Rnase L degrades viral and cellular RNAs, leading to inhibition of cellular protein synthesis and impairment of viral replication (reviewed in Ref. [62]). The proteins AIF1, IFI16, GBP2, MOP5, and IFI44 are involved in cell growth, cellular defence and cell proliferation. Note that the up-regulation of the IFN-γ-induced genes AIF1, IFI16, and GBP2 in the MRLN of LV-infected pigs at 3 dpi was not detected in pigs infected with the MV and HV strains. Interestingly, it has been reported that expression of AIF1 in vascular muscle cells is induced in response to arterial injury, a phenomenon also observed in CSFV pathogenesis [27], and IFI16 inhibits vascular tube morphogenesis and proliferation of primary endothelial cells [52].
Up-regulated genes involved in the innate/adaptive immune response include MHC1 genes such as SLA-1 and B2M, the natural killer surface receptor CD16b and genes such as immunoglobulins (J, lambda, kappa chains, heavy, light chains, FC fragment), which are ubiquitously expressed on the surface of B and T lymphocytes. The gene C4b was down-regulated. C4bp is a regulatory component (inhibitor) of the classical complement cascade [9] but also acts as an activator of B cells (by binding to CD40) [15, 49] and a protein that helps with the removal of apoptotic cells by binding to protein S [10, 68].
Up-regulated genes involved in ubiquitin-mediated proteolysis included a potential ubiquitine ligase (HERC6) and components of the proteasome (UBC and Psma6).
Most of the genes found to be down-regulated were structural components of the cytoskeleton (ACTA 2 and TPM1), genes involved in oxygen binding and transport (alpha–beta globin and ferroportin-1), or constituents of the mitochondrial respiratory chain (oxidative phosphorylation) (CytB, ND6, ND4L and E-NPP 3).
RT-PCR studies
Three up-regulated anti-viral genes (OAS2, CXCL10, TRAIL) and a gene (C4bp) that was down-regulated specifically in the MRNL were selected for real-time PCR studies. The expression levels of these genes were determined in all tissue sample collected from all individual pigs at 3, 7 and 10 dpi. In addition, IFNα expression and CSFV replication were also investigated in all these samples (Figs. 2, 3, 4). In agreement with the microarray data, a strong up-regulation of anti-viral gene expression was observed in all three immune tissues collected from LV-, MV- and HV-infected pigs at 3 dpi. At 7 and 10 dpi, this high level of expression dropped to lower levels, and in some pigs to the (normal) levels observed in uninfected pigs. In contrast, PCR results obtained for C4bp expression in MRLNs of LV- and HV-infected pigs at 3 dpi (Fig. 4) did not match with the data of the microarray, most likely due to a naturally occurring high inter-animal variation of C4bp expression in the MRLN (see control animals in Fig. 4). However, similar to what was observed for the MV strain, C4bp expression was significantly reduced at 7 and 10 dpi in MRLNs and spleens of most HV-infected pigs (Figs. 3, 4).
In all three immune organs of most MV- and HV-infected pigs, a high level of virus seemed to be associated with a low level of up-regulation of OAS2, CXCL10, and TRAIL. For LV-infected pigs, this inverse correlation between viral replication and anti-viral gene expression was also apparent in spleens and MRLNs obtained at 3 dpi and in spleens obtained at 7 dpi. However, in spleens and MLRNs collected from LV-infected pigs at 10 dpi, hardly any (or no) viral replication could be detected and no significant up-regulation of these anti-viral genes was observed. These results suggested that regulation of OAS2, CXCL10, and TRAIL gene expression in these immune organs is tightly linked to replication of CSFV. Interestingly, an inverse correlation between virus replication and IFNα gene expression was also observed in immune organs of a few infected pigs (marked with an asterisk in Fig. 4, p-value <0.05). Because IFNα gene expression in uninfected pigs was very capricious (see control animals in Fig. 4), more data are needed to draw conclusions about this.
Discussion
In the present study, we investigated the factors involved in the immune response of pigs during CSFV infection by determining gene expression profiles in immune organs. We demonstrate the extent of the porcine immune response against CSFV and the modulation of the host-signalling pathways by the virus. Besides the up-regulation of a cluster of IFN-γ-induced genes (AIF-1, IFI16, and GBP-2) in MRLN specifically induced by low virulent Brescia, no other pronounced qualitative differences in modulation of host pathways were detected between the strains of different virulence tested here. This emphasizes the complex interplay between CSFV and its host, in which subtle differences in genetic background of the virus and subtle differences in host factors, e.g., age and breed [21, 48], are decisive for the course of this viral disease.
The gene expression profiles provided by microarray indicated that enty and replication of CSFV modulated every part of the porcine immune system: (1) innate immunity with its humoral and chemical barriers such as the complement system and inflammation, as well as cellular barriers such as phagocytes, and (2) adaptive immunity, which involves B cells, killer T cells, and helper T cells.
Cytokines and chemokines such as IL6, IL8, TNF-α, IFNs, IL10 and IL12 have been demonstrated to be induced by CSFV [7, 38]. However, in the present study, CXCL10 is the most strongly up-regulated cytokine. CXCL10 (alias IP10) has been reported to be involved in effector T cell, monocyte and NK recruitment in response to infections with viruses involved in neurological disease, liver disease, heart disease and hemorrhagic fever disease [1, 18, 41, 44, 45]. It also has a pro-apoptotic effect via the p53-mediated (intrinsic) apoptotic pathway [76], which, according to our gene expression profiles, was also found to be activated by CSFV. This suggests that CXCL10 plays an important role in innate immunity through the recruitment of immune cells but also as a potential pro-apoptotic factor in CSFV disease.
The activation of TRAIL by CSFV confirms the conclusions of previous studies showing that programmed cell death is involved in the mechanism of lymphopenia during CSF [19, 66] and extends the list of death ligands known to be used by CSFV (TNF-α and FAS). Indeed, TRAIL functions as an antiviral protein [13, 72] by inducing cell death in infected cells via the extrinsic pathway but can also induce immunosuppression. It has been demonstrated that viruses such as HIV, measles virus, and cytomegalovirus infect DCs and induce TRAIL and TNF-α expression on their surface [64, 71]. Such DCs cannot present antigens and prime T cells for the generation of virus-specific CTLs. Instead, they kill interacting T, B and NK cells via the FAS and TRAIL/death-receptor pathway. This suggests that TRAIL plays a key role in the induction of apoptosis in uninfected cells and is also an immunosuppressor in CSF.
CFSV, by its replication in host cells, induced ER stress and oxidative stress, which intrinsically trigger apoptosis. As a countermeasure, several anti-apoptotic genes may have been activated, such as MCL1 (a gene related to Bcl-2), glutathione metabolism genes, and the gene for the Ca2+ regulation/transport protein calbindin. These observations are in agreement with previous in vitro studies with bovine virus diarrhoea virus, in which Bcl-2 gene expression was up-regulated in MDBK cells infected with a noncytopathic strain and down-regulated in these cells when infected with a cytopathic strain [6, 40]. This Bcl-2 down-regulation resulted in a reduced production of the antioxidant glutathione. Other viruses such as African swine fever virus or Epstein-Barr virus are known to regulate apoptosis by the synthesis of a viral protein homologous to Bcl-2 ([54], reviewed in Ref. [22]). Our results suggest that such anti-apoptotic gene activation, which may afford significant protection against virus-induced cell death, also may play a role in the pathogenesis of CSFV. No genes involved in NO metabolism were found to be regulated by CSFV.
The results show IFN-response-gene activation, although no significant up-regulation of IFN genes was found. This could be explained either by very early and short IFN expression (before day 3) [35] or by a low level of IFN expression produced by plasmacytoid DCs, which were not inhibited by CSFV [2, 57, 67] and probably undetectable by microarray due to the relatively low concentration of these cells in whole tissue samples.
CSFV also affects the regulation of the complement system by shutting down the normal host complement inhibitor C4bp. Since the complement regulator proteins (C4bp, factor H and factor I) have been shown to be major targets for inhibition of complement by viruses [20, 25], this down-regulation could reflect a viral defence response. However, C4pb has been implicated in other functions such as B-cell activation and the removal of apoptotic cells and bacteria (reviewed in Ref. [10]). Therefore, further investigation is necessary for a better comprehension of the C4bp function in CSF.
Ubiquitin-mediated proteolysis by the proteasome, another pathway identified by the microarray, is involved in directing protein trafficking and degradation (reviewed in Ref. [46]). In CSFV infection, the proteasome has been shown to degrade interferon regulatory factor 3 (IRF3) after interaction with viral protein N-Pro [4, 60], leading to the interruption of the transcription of IFNs. In addition, it has been shown that various other viruses can stimulate ubiquitin-mediated degradation of regulatory factors for their own benefit, for example, to support evasion of the host immune system (e.g., MHCI or STAT1 and 2, regulators of IFN expression; [26]), and to suppress apoptosis (e.g., the degradation of IAP, NFKB or Bcl-2; reviewed in Ref. [39]). Given that CFSV infection leads to immune suppression and apoptosis, the ubiquitin-proteasome is more likely to be used by CSFV to control, in addition of the IRF3, some of those crucial pathways. Anti-viral gene expressions (OAS2, CXCL10) and pro-apoptotic gene expression (TRAIL) showed a marked decrease when the virus replicated to a high level.
Although the WBC count is low in the animals at that stage of the disease (7 and 10 dpi), the depletion of T cells and DC cells does not by itself explain the decrease in gene expression. All of the gene data were normalized with 18S and beta-actin genes in order to eliminate this type of error. In addition, not all of the tested genes showed a decrease in gene expression. For example, MOP5 (data not shown) which is also an IFN-induced gene, showed no significant decrease in gene expression at 7 and 10 dpi.
This study also suggests that CSFV can modulate host-signalling pathways to enhance its persistence and survival.The exact mechanism of the modulation remains to be elucidated. However, several strategies have already been identified to control those pathways. For example, poliovirus, reovirus and influenza A virus can inhibit OAS activation by viral dsRNA-protein binding (reviewed in Ref. [63]). Several RNA and DNA viruses block IFN signaling in order to inhibit multiple pathways simultaneously, such as cellular antiviral enzymes (OAS, PKR, Mx1), anti-inflammatory response (CXCL10), and death ligands (TRAIL, FAS, TNF-α; reviewed in Ref. [53]). In addition to modulation of IFN production by Npro and Erns [28, 34, 37, 47], the pestivirus BVDV shows modulation of IFN signaling, such as PKR and MX activities, to establish a persistent infection [29, 59].
In conclusion, the functional analyses of gene expression show that most of the expressed genes contribute to anti-virus defense responses and pathogenesis of CSFV, while others reflect viral defense responses. Therefore, our findings can provide new insights into understanding the molecular basis of pathogenesis in CSFV pigs.
References
Arai K, Liu ZX, Lane T, Dennert G (2002) IP-10 and Mig facilitate accumulation of T cells in the virus-infected liver. Cell Immunol 219:48–56
Balmelli C, Vincent IE, Rau H, Guzylack-Piriou L, McCullough K, Summerfield A (2005) Fc gamma RII-dependent sensitisation of natural interferon-producing cells for viral infection and interferon-alpha responses. Eur J Immunol 35:2406–2415
Bauhofer O, Summerfield A, McCullough KC, Ruggli N (2005) Role of double-stranded RNA and Npro of classical swine fever virus in the activation of monocyte-derived dendritic cells. Virology 343:93–105
Bauhofer O, Summerfield A, Sakoda Y, Tratschin JD, Hofmann MA, Ruggli N (2007) Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J Virol 81:3087–3096
Bellido T, Huening M, Raval-Pandya M, Manolagas SC, Christakos S (2000) Calbindin-D28k is expressed in osteoblastic cells and suppresses their apoptosis by inhibiting caspase-3 activity. J Biol Chem 275:26328–26332
Bendfeldt S, Grummer B, Greiser-Wilke I (2003) No caspase activation but overexpression of Bcl-2 in bovine cells infected with noncytopathic bovine virus diarrhoea virus. Vet Microbiol 96:313–326
Bensaude E, Turner JL, Wakeley PR, Sweetman DA, Pardieu C, Drew TW, Wileman T, Powell PP (2004) Classical swine fever virus induces proinflammatory cytokines and tissue factor expression and inhibits apoptosis and interferon synthesis during the establishment of long-term infection of porcine vascular endothelial cells. J Gen Virol 85:1029–1037
Bidere N, Briet M, Durrbach A, Dumont C, Feldmann J, Charpentier B, de Saint-Basile G, Senik A (2002) Selective inhibition of dipeptidyl peptidase I, not caspases, prevents the partial processing of procaspase-3 in CD3-activated human CD8(+) T lymphocytes. J Biol Chem 277:32339–32347
Blom AM, Villoutreix BO, Dahlback B (2003) Mutations in alpha-chain of C4BP that selectively affect its factor I cofactor function. J Biol Chem 278:43437–43442
Blom AM, Villoutreix BO, Dahlback B (2004) Complement inhibitor C4b-binding protein-friend or foe in the innate immune system? Mol Immunol 40:1333–1346
Bots M, Medema JP (2006) Granzymes at a glance. J Cell Sci 119:5011–5014
Bouma A, de Smit AJ, de Kluijver EP, Terpstra C, Moormann RJ (1999) Efficacy and stability of a subunit vaccine based on glycoprotein E2 of classical swine fever virus. Vet Microbiol 66:101–114
Bouralexis S, Findlay DM, Evdokiou A (2005) Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis 10:35–51
Boyce M, Yuan J (2006) Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ 13:363–373
Brodeur SR, Angelini F, Bacharier LB, Blom AM, Mizoguchi E, Fujiwara H, Plebani A, Notarangelo LD, Dahlback B, Tsitsikov E, Geha RS (2003) C4b-binding protein (C4BP) activates B cells through the CD40 receptor. Immunity 18:837–848
Burkle A (2005) Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J 272:4576–4589
Carrasco CP, Rigden RC, Vincent IE, Balmelli C, Ceppi M, Bauhofer O, Tache V, Hjertner B, McNeilly F, van Gennip HG, McCullough KC, Summerfield A (2004) Interaction of classical swine fever virus with dendritic cells. J Gen Virol 85:1633–1641
Chen JP, Lu HL, Lai SL, Campanella GS, Sung JM, Lu MY, Wu-Hsieh BA, Lin YL, Lane TE, Luster AD, Liao F (2006) Dengue virus induces expression of CXC chemokine ligand 10/IFN-gamma-inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. J Immunol 177:3185–3192
Choi C, Hwang KK, Chae C (2004) Classical swine fever virus induces tumor necrosis factor-alpha and lymphocyte apoptosis. Arch Virol 149:875–889
Cummings KL, Waggoner SN, Tacke R, Hahn YS (2007) Role of complement in immune regulation and its exploitation by virus. Viral Immunol 20:505–524
Depner KR, Hinrichs U, Bickhardt K, Greiser-Wilke I, Pohlenz J, Moennig V, Liess B (1997) Influence of breed-related factors on the course of classical swine fever virus infection. Vet Rec 140:506–507
Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, Liverani E, Nix R, Silk R, Zhang F (2004) African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol 100:117–134
Doceul V, Charleston B, Crooke H, Reid E, Powell PP, Seago J (2008) The Npro product of classical swine fever virus interacts with IκBα, the NFκB inhibitor. J Gen Virol 89:1881–1889
Fahrenkrug SC, Smith TP, Freking BA, Cho J, White J, Vallet J, Wise T, Rohrer G, Pertea G, Sultana R, Quackenbush J, Keele JW (2002) Porcine gene discovery by normalized cDNA-library sequencing and EST cluster assembly. Mamm Genome 13:475–478
Favoreel HW, Van de Walle GR, Nauwynck HJ, Pensaert MB (2003) Virus complement evasion strategies. J Gen Virol 84:1–15
Gao G, Luo H (2006) The ubiquitin-proteasome pathway in viral infections. Can J Physiol Pharmacol 84:5–14
Gierens H, Nauck M, Roth M, Schinker R, Schurmann C, Scharnagl H, Neuhaus G, Wieland H, Marz W (2000) Interleukin-6 stimulates LDL receptor gene expression via activation of sterol-responsive and Sp1 binding elements. Arterioscler Thromb Vasc Biol 20:1777–1783
Gil LH, Ansari IH, Vassilev V, Liang D, Lai VC, Zhong W, Hong Z, Dubovi EJ, Donis RO (2006) The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J Virol 80:900–911
Gil LH, van Olphen AL, Mittal SK, Donis RO (2006) Modulation of PKR activity in cells infected by bovine viral diarrhea virus. Virus Res 116:69–77
Goodbourn S, Didcock L, Randall RE (2000) Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol 81:2341–2364
Gross G, van der Meulen J, Snel J, van der Meer R, Kleerebezem M, Niewold TA, Hulst MM, Smits MA (2008) Mannose-specific interaction of Lactobacillus plantarum with porcine jejunal epithelium. FEMS Immunol Med Microbiol 54:215–223
Haddad JJ (2004) Redox and oxidant-mediated regulation of apoptosis signaling pathways: immuno-pharmaco-redox conception of oxidative siege versus cell death commitment. Int Immunopharmacol 4:475–493
Heeres JT, Hergenrother PJ (2007) Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol 11:644–653
Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S (2006) The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol 80:11723–11732
Hulst M, Kerstens H, de Wit A, Smits M, van der Meulen J, Niewold T (2008) Early transcriptional response in the jejunum of germ-free piglets after oral infection with virulent rotavirus. Arch Virol 153:1311–1322
Hulst MM, van Gennip HG, Moormann RJ (2000) Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein E(rns). J Virol 74:9553–9561
Iqbal M, Poole E, Goodbourn S, McCauley JW (2004) Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J Virol 78:136–145
Jamin A, Gorin S, Cariolet R, Le Potier MF, Kuntz-Simon G (2008) Classical swine fever virus induces activation of plasmacytoid and conventional dendritic cells in tonsil, blood, and spleen of infected pigs. Vet Res 39:7
Jesenberger V, Jentsch S (2002) Deadly encounter: ubiquitin meets apoptosis. Nat Rev Mol Cell Biol 3:112–121
Jordan R, Wang L, Graczyk TM, Block TM, Romano PR (2002) Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J Virol 76:9588–9599
Kakimi K, Lane TE, Chisari FV, Guidotti LG (2001) Cutting edge: inhibition of hepatitis B virus replication by activated NK T cells does not require inflammatory cell recruitment to the liver. J Immunol 167:6701–6705
La Rocca SA, Herbert RJ, Crooke H, Drew TW, Wileman TE, Powell PP (2005) Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J Virol 79:7239–7247
Lee YJ, Chen JC, Amoscato AA, Bennouna J, Spitz DR, Suntharalingam M, Rhee JG (2001) Protective role of Bcl2 in metabolic oxidative stress-induced cell death. J Cell Sci 114:677–684
Liu MT, Chen BP, Oertel P, Buchmeier MJ, Armstrong D, Hamilton TA, Lane TE (2000) The T cell chemoattractant IFN-inducible protein 10 is essential in host defense against viral-induced neurologic disease. J Immunol 165:2327–2330
Liu MT, Keirstead HS, Lane TE (2001) Neutralization of the chemokine CXCL10 reduces inflammatory cell invasion and demyelination and improves neurological function in a viral model of multiple sclerosis. J Immunol 167:4091–4097
Liu YC, Penninger J, Karin M (2005) Immunity by ubiquitylation: a reversible process of modification. Nat Rev Immunol 5:941–952
Magkouras I, Matzener P, Rumenapf T, Peterhans E, Schweizer M (2008) RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J Gen Virol 89:2501–2506
Moennig V (2000) Introduction to classical swine fever: virus, disease and control policy. Vet Microbiol 73:93–102
Morgan BP, Marchbank KJ, Longhi MP, Harris CL, Gallimore AM (2005) Complement: central to innate immunity and bridging to adaptive responses. Immunol Lett 97:171–179
Niewold TA, Kerstens HH, van der Meulen J, Smits MA, Hulst MM (2005) Development of a porcine small intestinal cDNA micro-array: characterization and functional analysis of the response to enterotoxigenic E. coli. Vet Immunol Immunopathol 105:317–329
Pham CT, Ley TJ (1999) Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA 96:8627–8632
Raffaella R, Gioia D, De Andrea M, Cappello P, Giovarelli M, Marconi P, Manservigi R, Gariglio M, Landolfo S (2004) The interferon-inducible IFI16 gene inhibits tube morphogenesis and proliferation of primary, but not HPV16 E6/E7-immortalized human endothelial cells. Exp Cell Res 293:331–345
Randall RE, Goodbourn S (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47
Revilla Y, Cebrian A, Baixeras E, Martinez C, Vinuela E, Salas ML (1997) Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domain. Virology 228:400–404
Ruggli N, Tratschin JD, Schweizer M, McCullough KC, Hofmann MA, Summerfield A (2003) Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of N(pro). J Virol 77:7645–7654
Ruggli N, Bird BH, Liu L, Bauhofer O, Tratschin JD, Hofmann MA (2005) N(pro) of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-alpha/beta induction. Virology 340:265–276
Ruggli N, Summerfield A, Fiebach AR, Guzylack-Piriou L, Bauhofer O, Lamm CG, Waltersperger S, Matsuno K, Liu L, Gerber M, Choi KH, Hofmann MA, Sakoda Y, Tratschin JD (2009) Classical swine fever virus can remain virulent after specific elimination of the interferon regulatory factor 3-degrading function of Npro. J Virol 83:817–829
Sato M, Mikami O, Kobayashi M, Nakajima Y (2000) Apoptosis in the lymphatic organs of piglets inoculated with classical swine fever virus. Vet Microbiol 75:1–9
Schweizer M, Matzener P, Pfaffen G, Stalder H, Peterhans E (2006) “Self” and “nonself” manipulation of interferon defense during persistent infection: bovine viral diarrhea virus resists alpha/beta interferon without blocking antiviral activity against unrelated viruses replicating in its host cells. J Virol 80:6926–6935
Seago J, Hilton L, Reid E, Doceul V, Jeyatheesan J, Moganeradj K, McCauley J, Charleston B, Goodbourn S (2007) The Npro product of classical swine fever virus and bovine viral diarrhea virus uses a conserved mechanism to target interferon regulatory factor-3. J Gen Virol 88:3002–3006
Silverman RH (1997) 2–5A-dependent RNase L: a regulated endoribonuclease in the interferon system. In: Riordan GDAJF (ed) Ribonucleases: structure and function. Academic Press, New York, pp 515–551
Silverman RH, Cirino NM (1997) RNA decay by the interferon-regulated 2–5A system as a host defense against viruses. In: Morris JBHDR (ed) mRNA metabolism and post-transcriptional gene regulation. Wiley, New York, pp 295–309
Silverman RH (2007) Viral encounters with 2′, 5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729
Strater J, Moller P (2004) TRAIL and viral infection. Vitam Horm 67:257–274
Summerfield A, Hofmann MA, McCullough KC (1998) Low density blood granulocytic cells induced during classical swine fever are targets for virus infection. Vet Immunol Immunopathol 63:289–301
Summerfield A, Knotig SM, McCullough KC (1998) Lymphocyte apoptosis during classical swine fever: implication of activation-induced cell death. J Virol 72:1853–1861
Summerfield A, Alves M, Ruggli N, de Bruin MG, McCullough KC (2006) High IFN-alpha responses associated with depletion of lymphocytes and natural IFN-producing cells during classical swine fever. J Interferon Cytokine Res 26:248–255
Trouw LA, Bengtsson AA, Gelderman KA, Dahlback B, Sturfelt G, Blom AM (2007) C4b-binding protein and factor H compensate for the loss of membrane-bound complement inhibitors to protect apoptotic cells against excessive complement attack. J Biol Chem 282:28540–28548
Van Gennip HG, Vlot AC, Hulst MM, De Smit AJ, Moormann RJ (2004) Determinants of virulence of classical swine fever virus strain Brescia. J Virol 78:8812–8823
van Rijn PA, Wellenberg GJ, Hakze-van der Honing R, Jacobs L, Moonen PL, Feitsma H (2004) Detection of economically important viruses in boar semen by quantitative RealTime PCR technology. J Virol Methods 120:151–160
Vidalain PO, Azocar O, Lamouille B, Astier A, Rabourdin-Combe C, Servet-Delprat C (2000) Measles virus induces functional TRAIL production by human dendritic cells. J Virol 74:556–559
Warke RV, Martin KJ, Giaya K, Shaw SK, Rothman AL, Bosch I (2008) TRAIL is a novel antiviral protein against dengue virus. J Virol 82:555–564
Xu X, Fu XY, Plate J, Chong AS (1998) IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res 58:2832–2837
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15
Zaffuto KM, Piccone ME, Burrage TG, Balinsky CA, Risatti GR, Borca MV, Holinka LG, Rock DL, Afonso CL (2007) Classical swine fever virus inhibits nitric oxide production in infected macrophages. J Gen Virol 88:3007–3012
Zhang HM, Yuan J, Cheung P, Chau D, Wong BW, McManus BM, Yang D (2005) Gamma interferon-inducible protein 10 induces HeLa cell apoptosis through a p53-dependent pathway initiated by suppression of human papillomavirus type 18 E6 and E7 expression. Mol Cell Biol 25:6247–6258
Acknowledgments
This research was partly financed by the Dutch Ministry of Agriculture, Nature and Food Quality (WOT-01-003-010-01 and WOT-01-003-032-01). The work was additionally supported by the European Union through the network of excellence “EPIZONE”.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Durand, S.V.M., Hulst, M.M., de Wit, A.A.C. et al. Activation and modulation of antiviral and apoptotic genes in pigs infected with classical swine fever viruses of high, moderate or low virulence. Arch Virol 154, 1417–1431 (2009). https://doi.org/10.1007/s00705-009-0460-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0460-3