Abstract
The order Picornavirales includes several plant viruses that are currently classified into the families Comoviridae (genera Comovirus, Fabavirus and Nepovirus) and Sequiviridae (genera Sequivirus and Waikavirus) and into the unassigned genera Cheravirus and Sadwavirus. These viruses share properties in common with other picornavirales (particle structure, positive-strand RNA genome with a polyprotein expression strategy, a common replication block including type III helicase, a 3C-like cysteine proteinase and type I RNA-dependent RNA polymerase). However, they also share unique properties that distinguish them from other picornavirales. They infect plants and use specialized proteins or protein domains to move through their host. In phylogenetic analysis based on their replication proteins, these viruses form a separate distinct lineage within the picornavirales branch. To recognize these common properties at the taxonomic level, we propose to create a new family termed “Secoviridae” to include the genera Comovirus, Fabavirus, Nepovirus, Cheravirus, Sadwavirus, Sequivirus and Waikavirus. Two newly discovered plant viruses share common properties with members of the proposed family Secoviridae but have distinct specific genomic organizations. In phylogenetic reconstructions, they form a separate sub-branch within the Secoviridae lineage. We propose to create a new genus termed Torradovirus (type species, Tomato torrado virus) and to assign this genus to the proposed family Secoviridae.
Similar content being viewed by others
Introduction
The creation of the order Picornavirales has given a useful framework for the taxonomy of a large group of viruses, previously referred to as ‘picorna-like viruses’ or ‘members of the picornavirus-like superfamily or supergroup’ [23]. Members of the order Picornavirales share many common properties. They all have small icosahedral particles (25–30 nm) with a pseudo-T = 3 symmetry. The coat protein(s) structure is similar among picornavirales. It is made up of jelly rolls that can all be present in one large CP, or divided among two or three smaller CPs [8, 23 and references therein]. All picornavirales have a positive-strand RNA genome that may be monopartite or bipartite. Each genome segment encodes a large polyprotein. This polyprotein is cleaved by cysteine proteinases (such as the picornavirus 3C proteinase) that are structurally related to chymotrypsin [1, 9, 13]. The genome organization is conserved among picornavirales. The genome contains a typical ‘replication block’ that includes a type III helicase, the 3C-like proteinase and a type I RNA-dependent RNA-polymerase (Hel–Pro–Pol) [23]. The order Picornavirales includes viruses that infect vertebrates, insects, arthropods, higher plants, fungi and algae.
The family Comoviridae was for a long time the only known group of plant “picorna-like viruses” [12, 18]. Members of the family Comoviridae have bipartite genomes and contain either a single coat protein (genus Nepovirus) or two coat proteins (genera: Comovirus and Fabavirus that are mainly distinguished on the basis of their mode of transmission and hierarchical clustering of amino acid sequences). The discovery of plant “picorna-like viruses” with a monopartite genome and three CP subunits resulted in the creation of a distinct family, Sequiviridae, which currently includes two genera (Sequivirus and Waikavirus, distinguished by their specific genomic organization and mode of transmission) [19]. The molecular characterization of a number of new viruses as well as viruses previously considered to be members of the genus Nepovirus revealed that they differ significantly from nepoviruses, comoviruses and fabaviruses [22]. These new viruses have a bipartite genome and encode either two or three CP subunits, which distinguishes them from the nepoviruses. In phylogenetic studies based on their replication proteins, they were found to be related to members of the families Comoviridae or Sequiviridae, but branched in separate clades. Two new genera were created [22]. The genus Cheravirus includes viruses that have three CPs [20], and the genus Sadwavirus includes viruses that have two CPs [21]. Originally, various studies proposed that the new genera should be included in the families Comoviridae or Sequiviridae or that a new family should be created [10, 14, 16, 17, 24, 40]. However, at the time of creation of these genera, the executive committee from the International Committee for the Taxonomy of Viruses (ICTV) felt that a reevaluation of the taxonomy of plant picorna-like viruses was necessary before the two new genera could be assigned to either existing families or to a new family. Thus, for the time being, the genera Cheravirus and Sadwavirus are included in the order Picornavirales but are not assigned to any family [20–23].
The current classification has limitations in that properties shared by cheraviruses, sadwaviruses and other plant viruses classified in the order Picornavirales are not recognized. In addition, some viruses are still difficult to assign to existing genera/families. For example, strawberry latent ringspot virus (SLRSV), a founder member of the nepovirus “group” is currently considered a tentative sadwavirus because it encodes two CP subunits, but it clusters with cheraviruses in phylogenetic analysis using the amino acid sequence of replication proteins and has other characteristics typical of cheraviruses [22]. Recently, the elucidation of the complete genomic sequence of tomato torrado virus (ToTV) and tomato marchitez virus (ToMarV), two plant “picorna-like viruses” of a new type, also suggests that they do not belong to any of the existing genera and that a new genus should be created within the order Picornavirales [44, 45]. The “plant picornavirales” ICTV study group reevaluated the taxonomy of this group of viruses and proposed to create a new family that would encompass all plant viruses that currently belong to the order Picornavirales (these viruses will be referred to as plant picornavirales in the remainder of this paper). Because the proposed family is mainly an amalgamation of the families Sequiviridae and Comoviridae (as well as the unassigned genera Cheravirus and Sadwavirus), the name “Secoviridae” was selected for the new family. The family is proposed to include the genera Comovirus, Fabavirus, Nepovirus, Sequivirus, Waikavirus, Cheravirus and Sadwavirus (Table 1). In addition, the study group proposed to create a new genus termed Torradovirus and to assign this new genus to the proposed family Secoviridae. The ICTV executive committee approved the proposals in 2008 and posted them for ratification by the ICTV membership. This paper discusses the rationale for the creation of the family Secoviridae and genus Torradovirus and outlines the common characteristics of members of the new family as well as criteria distinguishing genera within the family.
Rationale for the creation of the proposed family Secoviridae
Although the diversity in the genomic organization of plant picornavirales is apparent (Fig. 1), these viruses share many common features. In addition to having characteristics in common with other picornavirales as discussed above (general capsid structure, polyprotein strategy for expression of proteins, conserved Hel–Pro–Pol replication block), plant picornavirales share distinctive features that differentiate them from other picornavirales.
Genome organization of plant picornavirales. Hel Helicase, Pro 3C-like Proteinase, Pol RNA-dependent RNA polymerase, CP capsid protein, MP movement protein, An poly(A). Question marks (?) after MP or CP indicate that the position and/or function of the predicted protein is tentative. The open reading frames are shown by the boxes. Proteinase cleavage sites identified experimentally or deduced by sequence comparison are shown by the solid or dotted vertical lines, respectively. The VPg is shown with a circle (open circle: no experimental evidence for a VPg; a sequence homologous to the VPg of comoviruses is found in the RNA1 of fabaviruses). The RNA2 of comoviruses (and possibly fabaviruses) encodes two polyproteins differing by their translation initiation site as indicated by the second AUG above the CPMV polyprotein. Dashed lines below the 3′ or 5′ UTR represent areas of sequence identity between RNA1 and RNA2. Similarly, the shaded areas in the N-terminus region of the subgroup C nepovirus and sadwavirus polyproteins represent areas of sequence identity. Possible subgenomic RNAs are represented by the arrows below the waikavirus 3′ UTR. Virus acronyms are defined in Tables 1, 2 and 3 when not defined here: arabis mosaic virus (ArMV), tomato black ring virus (TBRV), tomato ringspot virus (ToRSV), apple latent spherical virus (ALSV)
Biological characteristics
All viruses considered in this paper infect plants. To successfully invade the plant, they use a specialized protein(s) (termed movement protein) and/or an adapted capsid protein(s) that facilitates their cell-to-cell and long-distance movement. Many plant picornavirales induce similar cytopathological effects in their hosts. The formation of tubular structures that traverse the cell wall and contain virus-like particles has been observed in cells infected by comoviruses, fabaviruses, nepoviruses and several definite or tentative members of the genera Cheravirus, Sadwavirus and Sequivirus [28, 30, 33, 34, 43]. In the case of comoviruses, nepoviruses and cheraviruses, experimental evidence has been provided indicating that the tubular structures enable cell-to-cell movement of the virus in the plant and that the viral movement protein is a structural component of the tubules [31, 32, 46, 48, 49]. The mechanism of cell-to-cell movement has not yet been elucidated for other viruses. One sequivirus (parsnip yellow fleck virus, PYFV) was reported to induce tubules that traverse the cell wall and contain virus-like particles [28]. Although these tubular structures are similar to those observed for comoviruses, nepoviruses and cheraviruses, it is not known whether they are required for cell-to-cell movement of the virus. A movement protein has not (yet) been identified in the genome of sequiviruses or waikaviruses.
Viruses must also spread from plant to plant. While many plant picornavirales use a specialized vector (aphid, beetle, whitefly, nematode or mite) to do so (Table 2), others do not apparently require a vector, and they disseminate via pollen or seed transmission. Thus, the requirement for a specific vector is not a distinctive feature of the family or even of a genus within the proposed family. For example, although nepoviruses have long been assigned to the genus based on their transmissibility by nematodes, at least one nepovirus (blackcurrant reversion virus, BRV) is transmitted by mites, one nepovirus (tobacco ringspot virus, TRSV) is transmitted by thrips and nematodes, and several other nepoviruses do not have apparent vectors [27, 34, 37]. In addition, several nematode-transmitted viruses previously considered as tentative or definite nepoviruses have now been reclassified as sadwaviruses or cheraviruses based on the number of capsid proteins they contain and their phylogenetic relationships [22].
Virus structure
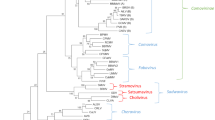
All plant picornavirales have icosahedral particles with a pseudo-T = 3 symmetry. The three jelly rolls, which are characteristic of picornavirales CP(s), can be present in one large CP with three jelly roll domains (Nepovirus), two CPs (one large CP including two jelly rolls and one smaller CP with a single jelly roll; Comovirus, Fabavirus and Sadwavirus) or three CPs (Sequivirus, Waikavirus, Cheravirus and the proposed new genus Torradovirus; see Table 2) [8, 23 and references therein]. While the number of CPs is currently considered a criterion for genus demarcation within the proposed family, this may need to be reevaluated. For example, SLRSV is currently considered a tentative sadwavirus because it has two CPs. However, the size of the two RNAs and the length of the 3′ untranslated regions are more similar to that of cheraviruses (Fig. 1). In addition, sequence comparisons reveal that SLRSV is more closely related to cheraviruses than to sadwaviruses (Fig. 2).
Hierarchical clustering of plant picornavirales based on the Pro–Pol amino acid sequence [23]. The families and genera are delineated on the right and the proposed new genus, sub-family and family are shown in the boxes. The letter in parenthesis after the nepovirus acronyms refers to the subgroup in which they have been classified (A, B or C). Circles indicate nodes supported by bootstrap values above 80% (closed circles) or 60% (open circles); nodes without circles are not supported to these levels. The bar represents a P distance of 0.1. The amino acid sequence clustering is based on the region between the Pro (CG) and the Pol (GDD) sequence motifs. The GenBank accession numbers used for each virus are as follows (virus acronyms are as defined in Tables 1, 2 and 3 or in Fig. 1, when not defined here): potato virus Y (PVY, NC 001616), Norwalk virus (NV, NC 001959), poliovirus (PV, NC 002058), Heterosigma akashiwo RNA virus (HaRNV, NC 005281), infectious flacherie virus (IFV, NC 003781), cricket paralysis virus (CrPV, NC 003924), PYFV (NC 003628), maize chlorotic dwarf virus (MCDV, NC 003626), RTSV (NC 001632), ToTV (NC 009013), ToMarV (NC 010987), SDV (NC 003785), SMoV (NC 003445), BRNV (NC 008182), SLRSV (NC 006964), stocky prune virus (StPV, AAZ76594), ALSV (NC 003787), CRLV (NC 006271), broad bean wilt virus 1 (BBWV1, NC 005289), BBWV2 (NC 003003), gentian mosaic virus (GeMV, BAD99001), squash mosaic virus (SqMV, NC 003799), radish mosaic virus (RaMV, AB295643), CPMV (NC 003549), red clover mottle virus (RCMV, NC 003741), bean pod mottle virus (BPMV, NC 003496), cowpea severe mosaic virus (CPSMV, NC 003545), raspberry ringspot virus (RpRSV, NC 005266), ToRSV (NC 003840), peach rosette mosaic virus (PRMV, AAB69867), cycas necrosis stunt virus (CNSV, NC 003791), grapevine chrome mosaic virus (GCMV, NC 003622), beet ringspot virus (BRSV, NC 003693), TBRV (NC 004439), ArMV (NC 006057), grapevine fanleaf virus (GFLV, NC 003615), TRSV (NC 005097), artichoke yellow ringspot virus (AYRSV, AM087671), BRV (NC 003509)
Genome organization and cleavage site specificity of the proteinase
Plant picornavirales have either a monopartite or bipartite genome. This could be an argument against the creation of a family regrouping these viruses. However, there are several other examples of families (e.g., Potyviridae, Closteroviridae) or even genera (e.g., Begomovirus in the family Geminiviridae) that include members that have either bipartite and monopartite genomes [2, 25, 36]. In spite of their apparent diversity, plant picornavirales share several similar features in their genomic organizations (Fig. 1). They all express their proteins as polyproteins and have a common Hel–Pro–Pol organization, a property shared with other picornavirales. In viruses that have a bipartite genome, the RNA2 polyprotein contains the CP domain(s) at its C-terminus, and the MP domain is always located immediately upstream of the CP domain. Information regarding the location of a putative MP domain is lacking for the viruses that have a monopartite genome.
In addition to the number of genomic RNA segments, there are other notable differences in the genome organization of plant picornavirales (Fig. 1), and these differences can be used as criteria for genus definition within the proposed family. First, the number of protein domains in the polyproteins, which are defined by the characterization of the cleavage sites recognized by the viral proteinase, can vary. For example, the presence of two domains upstream of the helicase domain on the RNA1-encoded polyprotein has been suggested to be a common feature of nepoviruses that distinguishes them from comoviruses or fabaviruses [47]. Second, the presence of additional open reading frames has been reported for members of the proposed genus Torradovirus [44] (see below) and for waikaviruses [11, 38], although additional studies will be required to confirm that these predicted ORFs are functional. Third, in the case of a waikavirus and a nepovirus, possible subgenomic RNAs that correspond to the long 3′ untranslated region have been detected in infected cells, although it is not clear whether these RNAs encode a protein and what their biological function may be [3, 35]. Fourth, although a polyA tail is present in most plant picornavirales, it is apparently absent from the genome of a definitive and a tentative sequivirus [15, 42], although it is present in another tentative member of this same genus [26]. Fifth, the cleavage site specificity of the proteinase varies (Table 2), sometimes even within a genus (e.g., Nepovirus).
Sequence comparisons
The conserved domains between the “CG” motif of the 3C proteinase and the “GDD” motif of the polymerase (Pro–Pol region) have been used to determine the relationships among picornavirales [23]. Phylogenetic studies using the Pro–Pol region reveal that all plant picornavirales group into a single branch when compared to other members of the order Picornavirales (see multiple sequence alignment generated with the ClustalX program [39] in Supplementary Fig. 1 and dendogram in Fig. 2). Although the level of diversity within this branch is high, it is in the same range as that currently accepted for the family Picornaviridae [23]. Within the plant picornavirales branch, several lineages are formed that generally represent the various genera within the proposed family Secoviridae (Fig. 2).
Defining characteristics of the proposed family Secoviridae and criteria defining genera
Considering the above, the study group proposed to create a new family termed Secoviridae to include the genera Comovirus, Fabavirus, Nepovirus, Sequivirus, Waikavirus, Cheravirus and Sadwavirus (Table 1). The study group also proposed to create the genus Torradovirus with the type species Tomato torrado virus and to include this genus in the family Secoviridae (Tables 1, 3). Finally, the study group considered the tight grouping of the currently recognized family Comoviridae (Fig. 2) and felt that this should be recognized at the taxonomic level. Consequently, it was suggested to create the subfamily Comovirinae within the family Secoviridae.
The criteria used to define genera within the proposed family will continue to be considered by the study group. At this time, we propose that some but perhaps not all criteria listed below may need to be met to define a genus. For example, members of the genus Nepovirus vary in the number of processing sites in their polyproteins (subgroup C nepoviruses have an additional protein domain on RNA-2) [7] and in the specificity of the proteinase [6], suggesting that this genus could be further divided (Fig. 1; Table 2). However, clustering in the Pro–Pol dendrogram does not support the current nepovirus subgroups (Fig. 2). The possibility that the genus Nepovirus should be split into several genera will be carefully considered by the study group in the coming year(s). Similarly, the genera Sadwavirus and Cheravirus will need to be further considered. The genus Cheravirus is well supported by the number of CPs, genomic organization and clustering in the Pro–Pol dendrogram [with the inclusion of Stocky prune virus (StPV) as a definite species, a proposal recently ratified by the ICTV membership] [5, 20, 22]. However, the genus Sadwavirus may need to be further defined. The genus is currently defined by the presence of two CPs [21]. As mentioned above, SLRSV is a tentative member of the genus because it encodes two CPs. However, it groups with cheraviruses in the Pro–Pol dendrogram and has a genomic organization similar to that of cheraviruses (Figs. 1, 2). The study group will therefore need to carefully consider the relationship of SLRSV to the two genera and may need to redefine the criteria for the two genera [22]. Similarly, the taxonomic position of strawberry mottle virus and black raspberry necrosis virus within the family Secoviridae will need to be redefined once more information is available regarding the genomic organization of RNA2 and the number of CPs (Table 2; Fig. 1) [22]. For the time being, we suggest that they should be considered tentative members of the genus Sadwavirus, based on their grouping in the Pro–Pol dendrogram (Fig. 2).
Finally, although the proposed subfamily Comovirinae is well supported by the Pro–Pol dendrogram, we felt that more information is required (as additional sequences become available) before other subfamilies can be suggested. For example, although the genera Sequivirus and Waikavirus share some common characteristics (number of RNAs and number of CPs), they also differ in other characteristics (presence or absence of a polyA tail and/or sub-genomic RNAs), and they do not branch together in the Pro–Pol dendrogram to the same level as that seen for the proposed subfamily Comovirinae (Fig. 2). The creation of other subfamilies within the family Secoviridae will be considered by the study group in the coming years along with the redefinition of some of the genera.
Common properties of viruses within the proposed family Secoviridae:
-
1.
Infect plants using specialized protein(s), so-called movement protein, and/or capsid protein(s) adapted for this function
-
2.
Common particle structure (icosahedral with pseudo-T = 3 symmetry)
-
3.
Positive-strand RNA genome that has a polyprotein expression strategy
-
4.
Common replication block including type III helicase, 3C-like cysteine proteinase, type I polymerase
-
5.
Clustering as a single branch in the Pro–Pol dendrogram when compared to members of the order picornavirales infecting other kingdoms
Examples of criteria defining genera within the proposed family Secoviridae:
-
1.
Number of genomic RNAs
-
2.
Number of protein domains and/or processing sites within the polyprotein(s)
-
3.
Number of CPs
-
4.
Presence of additional ORFs and/or subgenomic RNAs
-
5.
Clustering as a single branch in the Pro–Pol dendrogram when compared to other genera of the family Secoviridae
Not all criteria may need to be met simultaneously.
Rationale for the creation of the genus Torradovirus
Tomato torrado virus shares many characteristics with members of the proposed family Secoviridae, including similar virion structure and genome organization, with the typical Hel–Pro–Pol domains at the C-terminus of the RNA1-encoded polyprotein [44]. The RNA2-encoded polyprotein contains the domains for three CPs and a putative movement protein [44]. Phylogenetic studies using the Pro–Pol region reveal that ToTV branches with members of the proposed family Secoviridae when it is compared to other picornavirales [23]. A putative movement protein was identified in the genome of ToTV (the proposed type species for the new genus Torradovirus), but it has no homology with the movement proteins of members of the family Comoviridae, except for a very small motif (LxxPxL motif) [44]. It is not known whether ToTV or related viruses induce tubular structures to enable their movement from cell to cell.
The bipartite genome and the three CPs are properties that are shared by cheraviruses. However, other properties are unique to ToTV. In particular, a second partially overlapping reading frame was detected upstream of and partially overlapping with the polyprotein on RNA2 (Fig. 1). Recently, the complete sequence of ToMarV and the partial sequence of tomato apex necrosis virus became available [41, 45]. ToMarV and tomato apex necrosis virus are closely related to each other, suggesting that they are isolates of the same species [45]. A new isolate of ToTV (isolate Wal’03 from Poland) has recently been completely sequenced [4]. ToMarV shares many common properties with ToTV, including the presence of three CPs and an additional open reading frame on RNA2. In phylogenetic studies using the Pro–Pol region, a conserved region of the helicase domain or the CP domains, ToMarV, ToANV and the two ToTV isolates group together in a branch that is separated from other genera in the proposed family Secoviridae [4, 44, 45] (Fig. 2).
Tomato torrado virus was reported to be transmitted by whiteflies (Trialeurodes vaporariorum) [4, 29]. This vector has not been reported to be involved in transmission of other members of the proposed family Secoviridae. Information is not available regarding vector transmission of ToMarV or ToANV.
Although ToTV and ToMarV are related, the sequence identity at the amino acid level is 65% for the entire RNA1-encoded polyprotein and 78% in the Pro–Pol region. In the RNA2-encoded proteins of ToTV and ToMarV, the first open reading frames share 63% sequence identity (at the a.a. level), while the second open reading frames share 66% sequence identity. The a.a. sequence identity for the three CPs combined is 75%. In addition, the length of the 3′ untranslated regions differs between the two viruses and has less than 50% nucleotide sequence identity. Based on criteria currently accepted to distinguish species in other genera of the proposed family Secoviridae, this suggests that these two viruses constitute distinct species. In contrast, ToMarV and ToANV are highly similar (99% sequence identity for the Pro–Pol region and 89–98% for the CPs) [45], confirming that they are likely two isolates of the same species. Similarly, the two isolates of ToTV share a high degree of sequence identity (99–100% in the Pro–Pol and CPs) [4].
Based on this information, the study group proposed to create the genus Torradovirus within the new family Secoviridae and to assign Tomato torrado virus as the type species (Tables 1, 3). The term torradovirus is derived from tomato torrado virus. In Spanish, “torrado” means “toasted” to refer to the severe necrosis (burnt-like phenotype) observed in the disease induced by ToTV.
Example of criteria defining species within the proposed genus Torradovirus
-
1.
Less than 75% amino acid sequence identity in the CPs within a species
-
2.
Less than 80% amino acid sequence identity in the proteinase-polymerase region
-
3.
Type of biological vector
-
4.
Host range
-
5.
Absence of serological cross-reaction
-
6.
Absence of cross-protection
These criteria were defined by analogy with criteria established for other known genera within the proposed family Secoviridae. They may need to be adjusted as more viruses become characterized. Not all criteria may need to be met simultaneously.
References
Bazan JF, Fletterick RJ (1988) Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci USA 85:7872–7876
Berger PH, Adams MJ, Barnett OW, Brunt AA, Hammond J, Hill JH, Jordan RI, Kashiwazaki S, Rybicki E, Spence N, Stenger DC, Ohki ST, Uyeda I, van Zaayen A, Valkonen J, Vetten HJ (2005) Potyviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eighth report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 819–841
Brooks M, Bruening G (1995) A subgenomic RNA associated with cherry leafroll virus infections. Virology 211:33–41
Budziszewska M, Obrepalska-Steplowska A, Wieczorek P, Pospieszny H (2008) The nucleotide sequence of a Polish isolate of Tomato torrado virus. Virus Genes 37:400–406
Candresse T, Svanella-Dumas L, Le Gall O (2006) Characterization and partial genome sequence of stocky prune virus, a new member of the genus Cheravirus. Arch Virol 151:1179–1188
Carrier K, Hans F, Sanfacon H (1999) Mutagenesis of amino acids at two tomato ringspot nepovirus cleavage sites: effect on proteolytic processing in cis and in trans by the 3C-like protease. Virology 258:161–175
Carrier K, Xiang Y, Sanfacon H (2001) Genomic organization of RNA2 of Tomato ringspot virus: processing at a third cleavage site in the N-terminal region of the polyprotein in vitro. J Gen Virol 82:1785–1790
Chandrasekar V, Johnson JE (1998) The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily. Structure 6:157–171
Dougherty WG, Semler BL (1993) Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev 57:781–822
Everett KR, Milne KS, Forster RL (1994) Nucleotide sequence of the coat protein genes of strawberry latent ringspot virus: lack of homology to the nepoviruses and comoviruses. J Gen Virol 75:1821–1825
Firth AE, Atkins JF (2008) Bioinformatic analysis suggests that a conserved ORF in the waikaviruses encodes an overlapping gene. Arch Virol 153:1379–1383
Goldbach R (1987) Genome similarities between plant and animal RNA viruses. Microbiol Sci 4:197–202
Gorbalenya AE, Donchenko AP, Blinov VM, Koonin EV (1989) Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett 243:103–114
Iwanami T, Kondo Y, Karasev AV (1999) Nucleotide sequences and taxonomy of satsuma dwarf virus. J Gen Virol 80:793–797
Jadao AS, Krause-Sakate R, Liberti D, Pavan MA, Echer MM, Svanella-Dumas L, Zerbini FM, Candresse T, Le Gall O (2007) Further characterization of two sequiviruses infecting lettuce and development of specific RT-PCR primers. Arch Virol 152:999–1007
James D, Upton C (2002) Nucleotide sequence analysis of RNA-2 of a flat apple isolate of Cherry rasp leaf virus with regions showing greater identity to animal picornaviruses than to related plant viruses. Arch Virol 147:1631–1641
Karasev AV, Han SS, Iwanami T (2001) Satsuma dwarf and related viruses belong to a new lineage of plant picorna-like viruses. Virus Genes 23:45–52
Le Gall O, Iwanami T, Jones AT, Lehto K, Sanfacon H, Wellink J, Wetzel T, Yoshikawa N (2005) Comoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eighth report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 807–818
Le Gall O, Iwanami T, Jones AT, Lehto K, Sanfacon H, Wellink J, Wetzel T, Yoshikawa N (2005) Sequiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eighth report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 793–798
Le Gall O, Iwanami T, Jones AT, Lehto K, Sanfacon H, Wellink J, Wetzel T, Yoshikawa N (2005) Cheravirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eighth report of the international committee on the taxonomy of Viruses. Elsevier/Academic Press, London, pp 803–805
Le Gall O, Iwanami T, Jones AT, Lehto K, Sanfacon H, Wellink J, Wetzel T, Yoshikawa N (2005) Sadwavirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eighth report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 799–802
Le Gall O, Sanfacon H, Ikegami M, Iwanami T, Jones T, Karasev A, Lehto K, Wellink J, Wetzel T, Yoshikawa N (2007) Cheravirus and Sadwavirus: two unassigned genera of plant positive-sense single-stranded RNA viruses formerly considered atypical members of the genus Nepovirus (family Comoviridae). Arch Virol 159:1767–1774
Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE (2008) Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol 153:715–727
Li C, Yoshikawa N, Takahashi T, Ito T, Yoshida K, Koganezawa H (2000) Nucleotide sequence and genome organization of apple latent spherical virus: a new virus classified into the family Comoviridae. J Gen Virol 81:541–547
Martelli GP, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RHA, Dolja VV, Falk BW, Gonsalves D, Hu JS, Jelkmann W, Karasev AV, Minafra A, Namba S, Vetten HJ, Wisler GC, Yoshikawa N (2005) Closteroviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eight report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 1077–1087
Menzel W, Vetten HJ (2008) Complete nucleotide sequence of an isolate of the Anthriscus strain of Parsnip yellow fleck virus. Arch Virol 153:2173–2175
Messieha M (1969) Transmission of tobacco ringspot virus by thrips. Phytopathology 59:943–945
Murant AF, Roberts IM, Hutcheson AM (1975) Effects of parsnip yellow fleck virus on plant cells. J Gen Virol 26:277–285
Pospieszny H, Borodynko N, Hasiow B, Obrepalska-Steplowska A, Budziszewska M (2007) Tomato torrado virus, a new virus transmitted by greenhouse whitefly (Trialeurodes vaporariorum) in Poland. In: 10th international plant virus epidemiology meeting, Hyderabad, India
Pouwels J, Carette JE, Van Lent J, Wellink J (2002) Cowpea mosaic virus: effects on host cell processes. Mol Plant Pathol 3:411–418
Pouwels J, van der Velden T, Willemse J, Borst JW, van Lent J, Bisseling T, Wellink J (2004) Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. J Gen Virol 85:3787–3796
Ritzenthaler C, Schmit A-C, Michler P, Stussi-Garaud C, Pinck L (1995) Grapevine fanleaf nepovirus P38 putative movement protein is located on tubules in vivo. Mol Plant–Microbe Interact 8:379–387
Roberts IM, Harrison BD (1970) Inclusion bodies and tubular structures in Chenopodium amaranticolor plants infected with strawberry latent ringspot virus. J Gen Virol 7:47–54
Sanfacon H (2008) Nepovirus. In: Mahy BWJ, Van Regenmortel MH (eds) Encyclopedia of virology, vol 3, 3rd edn. Elsevier, Oxford, pp 405–413
Shen P, Kaniewska M, Smith C, Beachy RN (1993) Nucleotide sequence and genomic organization of rice tungro spherical virus. Virology 193:621–630
Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC (2005) Geminiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy, eight report of the international committee on the taxonomy of viruses. Elsevier/Academic Press, London, pp 301–326
Susi P (2004) Black currant reversion virus, a mite-transmitted nepovirus. Mol Plant Pathol 5:167–173
Thole V, Hull R (1996) Rice tungro spherical virus: nucleotide sequence of the 3′ genomic half and studies on the two small 3′ open reading frames. Virus Genes 13:239–246
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thompson JR, Leone G, Lindner JL, Jelkmann W, Schoen CD (2002) Characterization and complete nucleotide sequence of strawberry mottle virus: a tentative member of a new family of bipartite plant picorna-like viruses. J Gen Virol 83:229–239
Turina M, Ricker MD, Lenzi R, Masenga V, Ciuffo M (2007) A severe disease of tomato in the Culiacan area (Sinaloa, Mexico) is cause by a new picorna-like viral species. Plant Dis 91:341–932
Turnbull-Ross AD, Mayo MA, Reavy B, Murant AF (1993) Sequence analysis of the parsnip yellow fleck virus polyprotein: evidence of affinities with picornaviruses. J Gen Virol 74:555–561
Usugi T, Saito Y (1979) Satsuma dwarf virus. CMI/AAB descriptions of plant viruses no. 208. http://www.dpvweb.net/dpv/showdpv.php?dpvno=208
Verbeek M, Dullemans AM, van den Heuvel JF, Maris PC, van der Vlugt RA (2007) Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato. Arch Virol 152:881–890
Verbeek M, Dullemans AM, van den Heuvel JF, Maris PC, van der Vlugt RA (2008) Tomato marchitez virus, a new plant picorna-like virus from tomato related to tomato torrado virus. Arch Virol 153:127–134
Wellink J, van Lent JW, Verver J, Sijen T, Goldbach RW, van Kammen A (1993) The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J Virol 67:3660–3664
Wetzel T, Chisholm J, Bassler A, Sanfacon H (2008) Characterization of proteinase cleavage sites in the N-terminal region of the RNA1-encoded polyprotein from Arabis mosaic virus (subgroup A nepovirus). Virology 375:159–169
Wieczorek A, Sanfacon H (1993) Characterization and subcellular localization of tomato ringspot nepovirus putative movement protein. Virology 194:734–742
Yoshikawa N, Okada K, Asamuma K, Watanabe K, Igarasi A, Li C, Isogai M (2006) A movement protein and three capsid proteins are all necessary for the cell-to-cell movement of apple latent spherical cheravirus. Arch Virol 151:837–848
Acknowledgments
We would like to thank Joan Chisholm for careful editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2009_367_MOESM1_ESM.pdf
Multiple sequence alignment of the Pro–Pol region of members of the proposed family secoviridae and of representative members of other families or genera in the order Picornavirales. The alignment was produced using the ClustalX program and was used to generate the dendogram presented in Fig. 2 (Supplementary Fig. 1 PDF 257 kb).
Rights and permissions
About this article
Cite this article
Sanfaçon, H., Wellink, J., Le Gall, O. et al. Secoviridae: a proposed family of plant viruses within the order Picornavirales that combines the families Sequiviridae and Comoviridae, the unassigned genera Cheravirus and Sadwavirus, and the proposed genus Torradovirus. Arch Virol 154, 899–907 (2009). https://doi.org/10.1007/s00705-009-0367-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0367-z