Abstract
We investigate the timing of diversification in allopolyploids of Nicotiana (Solanaceae) utilising sequence data of maternal and paternal origin to look for evidence of a lag phase during which diploidisation took place. Bayesian relaxed clock phylogenetic methods show recent allopolyploids are a result of several unique polyploidisation events, and older allopolyploid sections have undergone subsequent speciation at the polyploid level (i.e. a number of these polyploid species share a singular origin). The independently formed recent polyploid species in the genus all have mean age estimates below 1 million years ago (Ma). Nicotiana section Polydicliae (two species) evolved 1.5 Ma, N. section Repandae (four species) formed 4 Ma, and N. section Suaveolentes (~35 species) is about 6 million years old. A general trend of higher speciation rates in older polyploids is evident, but diversification dramatically increases at approximately 6 Ma (in section Suaveolentes). Nicotiana sect. Suaveolentes has spectacularly radiated to form 35 species in Australia and some Pacific islands following a lag phase of almost 6 million years. Species have filled new ecological niches and undergone extensive diploidisation (e.g. chromosome fusions bringing the ancestral allotetraploid number, n = 24, down to n = 15 and ribosomal loci numbers back to diploid condition). Considering the progenitors of Suaveolentes inhabit South America, this represents the colonisation of Australia by polyploids that have subsequently undergone a recent radiation into new environments. To our knowledge, this study is the first report of a substantial lag phase being investigated below the family level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyploidy in angiosperms
Numerous whole-genome duplication (WGD) or polyploidisation events have been identified in the evolutionary history of flowering plants, leading to the now well-established concept that all angiosperms are paleopolyploids [recently reviewed in Soltis et al. (2016) and Wendel (2015)]. Astonishing levels of duplication have been documented in some angiosperms; for example, it has been estimated that Gossypium has a ploidy level of 144x (Wendel 2015). Each round of polyploidy is followed by diploidisation [see Dodsworth et al. (2016a)], which involves gene loss (often preferentially from one progenitor, i.e. biased fractionation), chromosome reorganisation, chromosome fusion and overall genome size reduction (Wendel 2015). Thus, for example, Brassica species have only 8–10 pairs of chromosomes and appear to be functionally ‘diploid’ in terms of the numbers of structural (housekeeping) genes despite their paleopolyploid ancestry (they have an estimated ploidy level of 288x; Wendel 2015). The extensive literature on isozymes (Hamrick and Godt 1989; Soltis and Soltis 1990), which looks at a set of standard housekeeping (structural) loci, documents that most angiosperms exhibit little evidence of being paleopolyploid.
Studies of diversification rates across angiosperm phylogenetic trees suggest polyploidy may facilitate bursts of speciation. Often species-poor clades are sister to clades that have undergone WGD and subsequently formed species-rich clades (Soltis et al. 2009). However, mounting evidence suggests species radiations do not immediately follow polyploid events, but rather appear after a fairly substantial time lag. The ‘WGD radiation lag-time model’ was proposed by Schranz et al. (2012) and stated that increased rates of lineage diversification only become evident after a lag phase that is typically measured in millions of years. Using cases from five plant families (Asteraceae, Brassicaceae, Fabaceae, Poaceae and Solanaceae), Schranz et al. (2012) suggested that post-lag radiations are often correlated with changing environmental conditions and migration to new areas. Diversification rates are known to be highly variable across the angiosperm tree of life. A formal diversification rates analysis using stepwise AIC (Akaike Information Criterion) identified rapid radiations within other radiations, termed ‘nested radiations’ by Tank et al. (2015). Increases in net diversification rates tend to follow a lag phase post-polyploidisation, adding support to the previously proposed model (Schranz et al. 2012; Tank et al. 2015). It therefore seems probable that at the genomic level the progression of events associated with diploidisation is an important factor in understanding the lag phase (Dodsworth et al. 2016a).
Genomic studies have provided a wealth of information about the rounds of polyploidy and diploidisation that have occurred during angiosperm evolution, but some limitations are insurmountable when studying ancient paleopolyploids. One problem is that the diploid progenitors of angiosperm paleopolyploids are unknown due to the long timeframes involved, which limits studies comparing the advantages conferred by polyploidy. For example, the diploid progenitors of the well-studied Arabidopsis thaliana (Brassicaceae) remain unknown and are most likely extinct. We therefore investigate the lag phase in a group of angiosperms, Nicotiana (Solanaceae), in which diploid progenitors and their polyploid descendants are known in order to tease apart the complex factors involved.
Polyploidy in Nicotiana
Our study system is the well-characterised genus Nicotiana (Solanaceae), which comprises approximately 75 species, 48% of which are allotetraploids (36 species). Close relatives of the diploid progenitors are known for all these allotetraploids with the exception of the maternal progenitor of Suaveolentes (Clarkson et al. 2010; Kelly et al. 2013). Some allopolyploids are recent and have no close polyploid relatives, whereas others appear to be much older and numerous, the result of speciation at the polyploid level. Polyploidy and diploidisation have been the subject of study in the genus for around two decades: chromosome arm translocations (Lim et al. 2004), homoploid hybridisation (Clarkson et al. 2010), intergenic recombination (Kelly et al. 2010), concerted evolution (Kovarik et al. 2004), long-term diploidisation of genomic repeats (Clarkson et al. 2005; Dodsworth et al. 2016a), progenitor determination (Kelly et al. 2013), maternal genome donors (Clarkson et al. 2004), elimination of genomic repeats (Renny-Byfield et al. 2011), the phylogenetic signal in repeats (Dodsworth et al. 2015, 2016b), floral evolution (McCarthy et al. 2015), morphological character evolution (Marks et al. 2011a; McCarthy et al. 2016), biogeography (Ladiges et al. 2011) and genome size changes (Leitch et al. 2008). An earlier paper (Clarkson et al. 2005) focused on the timing of polyploid events for a subset of polyploids in Nicotiana, using nonparametric rate smoothing (NPRS), but recent advances in molecular clock analysis make another study of this subject in the genus as a whole timely.
Time-calibrated phylogenetic trees and Nicotiana
Establishment of a timeframe for the progression of post-polyploidisation events discussed above will inform our understanding of biogeography, ecological niche differentiation and morphological diversification in Nicotiana. We know from present-day species distribution data for the genus that some species (both diploids and allopolyploids) appear to have undergone long-distance dispersal events (Clarkson et al. 2010). A previous study of Nicotiana section Repandae (Clarkson et al. 2005) used sister pairs of species on volcanic islands of known age and the continent as calibration points in order to yield maximum ages for polyploidisation. Nicotiana cordifolia is endemic to the island of Masafuera (formed approximately 2.4 million years ago; Ma) and is sister to the mainland (Chilean) N. solanifolia. Nicotiana stocktonii and N. nesophila are endemic to the Revillagigedo Islands (formed approximately 1.1 Ma) and are sister to mainland (Mexican) N. repanda (Clarkson et al. 2005), which yielded a maximum age for N. sect. Repandae of 5 Ma. However, our 2005 study presented only a few dates relevant to species of section Repandae, with further dates yielded by this analysis quoted in our later publication (Leitch et al. 2008). It should be noted that no time-calibrated phylogenetic trees were ever published for Nicotiana as a whole from our previous study.
A phylogenetic analysis of all genera of Solanaceae calibrated with two fossil calibration points (Särkinen et al. 2013) has revealed age estimates for Nicotiana and related genera. However, dates within Nicotiana are problematic and unreliable in that study due to the low levels of variation and potential incongruence of the loci combined [see tree in Supplementary Data File two of Särkinen et al. (2013)]. The current study is based on three of our previously published DNA sequence datasets (Clarkson et al. 2004, 2010; Kelly et al. 2013). Our intention is to produce time-calibrated phylogenetic trees using both maternally (plastid DNA) and biparentally inherited (low copy nuclear) loci. The main questions under investigation are:
-
1.
Do both the maternally and paternally inherited loci yield similar ages for polyploids in Nicotiana? (i.e. evidence of consistency between datasets).
-
2.
Are the dates consistent with external geological dates? (e.g. volcanic islands of known age).
-
3.
Is there evidence of a lag phase following polyploidisation and preceding diversification?
-
4.
Can we correlate specific diploidisation processes with the different ages of polyploids?
Materials and Methods
Preparation of sequence matrices
In order to produce a robust, well-resolved and well-supported nuclear phylogenetic analysis, the sequences from two previous datasets, nuclear-encoded plastid-expressed glutamine synthetase (GS) from Clarkson et al. (2010) and leafy/floricaula (LFY) from Kelly et al. (2013), were combined. The DNA sequences used in both studies were produced from the same accession, thus reducing sampling error. The combined nuclear matrix required some taxon pruning largely of GS sequences because the published GS dataset had 94 terminals, whereas that for LFY had 70 terminals. A recombinant GS sequence was previously detected for the diploid N. acuminata (Clarkson et al. 2010), and so this taxon was removed from the matrix. This diploid is not a progenitor of any allopolyploids and therefore will not affect the dates yielded for allopolyploid groups. Allotetraploid N. arentsii is absent from the LFY dataset (due to PCR failure), and therefore, this species was entirely removed from the nuclear dataset. This step was taken rather than using missing data because time-calibrated analyses can be sensitive to missing data, causing errors in calculation of branch lengths. The accession labelled N. debneyi in GS and N. forsteri in LFY are based on the same DNA sample; the former name is now considered a synonym of the latter (Marks 2010).
The origin of the maternal progenitor of N. section Suaveolentes is complex (Kelly et al. 2013). Breaking GS into the two pieces indicated by recombination break points results in the maternal progenitor of N. sect. Suaveolentes being related to two diploid sections: GS 5′ falls with N. sect Petunioides, whereas GS 3′ with N. sect Trigonophyllae. LFY places them with N. sylvestris (for both ‘maternal’ and ‘paternal’ copies of the locus), ADH with Petunioides, nrITS with Alatae, WAXY with Alatae and plastid with Noctiflorae. Therefore, this diploid progenitor was probably a hybrid species at the homoploid (diploid) level (Kelly et al. 2013). In view of the uncertainty and incongruence detected in previous studies, we therefore decided to use only the paternal copy for each allotetraploid in the combined nuclear analysis. The maternal and paternal progenitors of each allopolyploid are known from previous studies (Clarkson et al. 2010; Kelly et al. 2013). This information was used to identify copies and then remove the maternal copies from all allopolyploids in the nuclear dataset.
Diploid section Undulatae was removed from the combined nuclear dataset due to strongly supported incongruence between the two nuclear regions (GS and LFY). In the GS dataset, Undulatae and Paniculatae form a well-supported clade with PP 1.00 (Clarkson et al. 2010), which is congruent with all other phylogenetic datasets (e.g. plastid, Clarkson et al. 2004). However, the LFY dataset places Undulatae sister to section Tomentosae (Kelly et al. 2013) with high support (PP 1.00). Only two species of Undulatae were present in the LFY dataset (N. undulata and N. thyrsiflora), and therefore, the loss of this section represents only a small loss in overall species number for the combined nuclear dataset.
The second data matrix used in this study was the plastid DNA matrix of (Clarkson et al. 2004). This dataset consists of the following five plastid regions: matK, ndhF, trnS-G spacer, trnL intron and trnL-F spacer. This data matrix has the broadest taxon sampling: 55 species of Nicotiana plus 24 species of tribe Anthocercideae (the sister taxon of Nicotiana) and four more divergent Solanaceae outgroup taxa. Two artificial hybrids (N. × sanderae and N. × digluta) were removed from the dataset because they are not relevant to this study. All sequences used in this study are available at EMBL (http://www.ebi.ac.uk/).
The nucleotide substitution models used for Bayesian relaxed clock analyses of the datasets were determined using MrModeltest v2.3 (Nylander 2008). The models selected for GS were: codon position 1 = HKY, codon positions 2 and 3 = K80, introns = GTR + Γ. Models identified for the LEAFY dataset were codon position 1, 3 and introns = GTR + Γ, codon position 2 = GTR. Models identified for the plastid dataset were: codon position 1 and 2 = GTR + I+Γ, codon position 3 and noncoding = GTR + Γ.
Bootstrap analyses
A bootstrap analysis under maximum parsimony was performed on the combined nuclear dataset to give an indication whether the two regions (GS and LFY) are congruent. Previous studies in the genus showed that parsimony bootstrap percentages are more sensitive than Bayesian posterior probabilities for the detection of conflicting signal in datasets (see discussion section in Clarkson et al. (2010)). Increased levels of support for the combined trees, in comparison with the single gene trees, indicate congruence, and decreased support levels indicate incongruence. Bootstrap analysis (Felsenstein 1985) was performed using PAUP version 4.0b (Swofford 2001) using the following parameters: tree-bisection-reconnection (TBR) branch swapping, simple taxon addition, 1000 bootstrap replicates and saving ten trees per replicate.
Bayesian relaxed clock analyses
The default Bayesian settings were used in BEAST2 version 2.4.1 (Bouckaert et al. 2014) unless otherwise stated (see below). The tree topology and node ages are estimated simultaneously in BEAST2, and therefore, phylogenetic uncertainty can be used to assess confidence levels. Highest posterior density (HPD) was used to assess the confidence in each estimated node age because this measure acts as a Bayesian analogue of confidence intervals. It should be stressed that independent analyses were run using the nuclear dataset and the plastid dataset throughout; these two datasets were never combined. Analyses were run using the following parameters: uncorrelated log-normal relaxed clock model (to account for rate variation across the different branches), substitution models were tailored to each dataset (see previous section for full list of data partitions), birth/death model tree prior used (this is designed to account for extinction in addition to speciation). Trees were calibrated with the Symonanthus–Nicotiana split of 15 Ma from Särkinen et al. (2013) as a secondary calibration point, using a normal distribution centred on 15 Ma with a standard deviation of one in order to reflect the uncertainty associated with the original estimate. The MCMC chain was run for 30 million generations for two independent runs. Analyses were performed on the CIPRES Science Gateway V.3.3 (http://www.phylo.org/). Convergence and burn-in were checked using Tracer v1.6 (Rambaut et al. 2014). A 10% burn-in was discarded, and Tree Annotator v1.8.2 (Rambaut and Drummond 2014) was used to find the maximum clade credibility (MCC) tree and to summarise the posterior samples of trees produced by BEAST2. Trees were viewed using FigTree v1.4.2 (Rambaut 2014).
Using the Symonanthus–Nicotiana 15 million years ago (Ma) calibration, the N. solanifolia–N. cordifolia maximum age that was predicted to be around 2.4 Ma based on geological data (Clarkson et al. 2005) can be evaluated. Similarly, the maximum age of the N. repanda–N. nesophila and N. stocktonii node that we predicted to be approximately 1.1 Ma based on geological data (Clarkson et al. 2005) can also be evaluated. A second pair of analyses (on the plastid and nuclear datasets), using the parameters outlined above, was performed using BEAST2 with the two geological calibration points only. This was performed to enable the cause of any age discrepancies to be identified such as differences due to the previously used NPRS method versus BEAST2 or fossil versus geological. However, it must be considered that the geological ages are maximum estimates and the fossil dates are minimum ages, and therefore, we might expect some degree of discrepancy.
The two independent age estimates, plastid and nuclear, were used to produce a combined mean age (in millions of years) for each polyploid group. Combined mean age was plotted against number of species for each polyploid group using an XY scatter plot in Excel (version 15.0.4875.1000).
Results
Dataset attributes
The combined nuclear matrix contained an aligned length of 2824 characters after areas of ambiguous alignment were excluded and 1066 characters (38%) were variable. The plastid matrix contained an aligned length of 4305 characters with 808 (19%) variable. The two alignments used (plastid matrix and nuclear matrix) are available at TreeBase (https://treebase.org/). The combined nuclear matrix yields a well-supported tree for the genus Nicotiana with many nodes receiving around 100% bootstrap (see Online Resource 1: Bootstrap Figure). This was a preliminary step to ensure we have a robust dataset for the subsequent time-calibrated analyses. The phylogenetic trees resulting from BEAST2 analyses of both the plastid and nuclear datasets were consistent with previous studies in the genus.
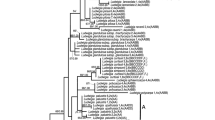
Using 15 Ma for the Symonanthus–Nicotiana split as a calibration point, these data yield an age for the N. solanifolia–N. cordifolia split at approximately 2.9 Ma for the plastid dataset (95% HPD 1.3–4.5) and c. 2.3 Ma for the nuclear dataset (95% HPD 1.1–3.2). This node was predicted to be approximately 2.4 Ma based on geological data (Clarkson et al. 2005). These data yield age estimates of c. 0.7 Ma for the plastid dataset (95% HPD 0.1–1.3) and c. 0.9 Ma for the nuclear dataset (95% HPD 0.5–1.4) for the N. repanda–N. nesophila/N. stocktonii node, which we would predict to be approximately 1.1 Ma based on geological data (Clarkson et al. 2005). These results therefore show that the datasets yield dates that are consistent with published ages based on geological dates. The results yielded by time-calibrated analyses are summarised in Fig. 1 (plastid tree) and Fig. 2 (nuclear tree).
Phylogenetic tree resulting from the Bayesian relaxed molecular clock method implemented in BEAST2 (Bouckaert et al. 2014). The plastid tree based on five loci (matK, ndhF, trnS-G spacer, trnL intron and trnL-F spacer) is shown above. The red clades indicate allotetraploids, and the grey bars show the 95% HPD interval of node ages
Phylogenetic tree resulting from the Bayesian relaxed molecular clock method implemented in BEAST2 (Bouckaert et al. 2014). The combined nuclear tree is shown, which is based on GS and LFY with only paternal copies included for allopolyploids. The dataset has all maternal copies removed due to the problems associated with the maternal copy of Suaveolentes (i.e. recombinant according to Kelly et al. (2013)). The red clades indicate allotetraploids, and the grey bars show the 95% HPD interval of node ages
Age estimates for allotetraploid sections
The nodes associated with divergence from the most recent common ancestor (MRCA) for each allopolyploid section (allotetraploid to diploid progenitor ages) are reported in this section. All dates presented are the mean age estimates, in millions of years, yielded by BEAST2 analyses (Figs. 1, 2). The progenitors of each allopolyploid were identified in previous studies (Clarkson et al. 2010; Kelly et al. 2013). Nicotiana sect. Suaveolentes is the oldest and most species-rich allotetraploid section with age estimates of c. 6.4 Ma from its maternal progenitor section Noctiflorae (95% HPD 4.9–8.0, Fig. 1) and c. 5.5 Ma from its paternal progenitor N. sylvestris (95% HPD 4.4–6.9, Fig. 2). Section Repandae contains four species including two island endemics, and the section yields age estimates of c. 5.1 Ma from its maternal progenitor N. sylvestris (95% HPD 3.2–7.0) and c. 3.5 Ma from its paternal progenitor N. obtusifolia (95% HPD 2.4–4.9). Polydicliae is the only allopolyploid section found in western North America with an age of c. 1.2 Ma from its maternal progenitor N. obtusifolia (95% HPD 0.2–2.2) and c. 1.5 Ma from its paternal progenitor N. attenuata (95% HPD 0.5–2.1). Nicotiana tabacum the tobacco of commerce yields an age estimate of c. 0.4 Ma from its maternal progenitor N. sylvestris (95% HPD 0.1–1.1) and c. 0.8 Ma from its paternal progenitor N. tomentosiformis (95% HPD 0.3–1.3). Nicotiana arentsii is an allotetraploid native to the Peruvian Andes with an age estimate of c. 0.4 Ma from its maternal progenitor N. undulata (95% HPD 0.1–1.0). Nicotiana rustica is an allotetraploid native to South America with an age estimate of c. 0.6 Ma from its maternal progenitor N. paniculata (95% HPD 0.2–1.2). In each case above, progenitor species are defined as the most closely related extant diploid species to the actual progenitors of the allotetraploids. Mean age estimates for all clades (both polyploid and diploid) are displayed in Online Resources 2 and 3—Mean Age Estimates.
Combining the mean age estimates yielded by the plastid and nuclear datasets produced a combined mean age for each polyploid group in millions of years (Table 1). The two datasets are generally consistent, but there are some differences particularly for the age of Repandae. When combined mean age is plotted against the number of species per polyploid group, a general trend of more diversification is evident over time. However, N. sect. Suaveolentes in particular has undergone a rapid radiation in the last six million years to form the species-rich group (35 species) of today (see Fig. 3).
Effect of polyploid age on speciation in Nicotiana. An XY scatter graph showing the number of species resulting from each polyploidy event plotted against the estimated age of the WGD. A lag phase is evident up to approximately 4–6 Ma followed by increased diversifications rates. The numbers shown above the data points denote the number of species derived from each WGD
Age estimates for diploid sections
The diploid ages are not the principal focus of this investigation, but are summarised here so that they can inform other studies if required. Dates for only the MRCA of each diploid section are provided (see datasets presented in Figs. 1 and 2). For a full list of species recognised in each diploid section, see Knapp et al. (2004). Nicotiana sect. Alatae comprises eight species (not all of which are included here), and the section yields age estimates of c. 6.2 Ma from the plastid analysis and c. 7.7 Ma from the nuclear analysis. Noctiflorae contains six species with age estimates of c. 7.2 Ma from the plastid analysis and c. 9.1 Ma from the nuclear analysis. Paniculatae comprises seven species, and the section yields age estimates of c. 7.7 Ma from the plastid analysis and c. 9.0 Ma from the nuclear analysis. Petunioides has eight species, and the section yields age estimates of c. 9.8 Ma from the plastid analysis and c. 9.1 Ma from the nuclear analysis. Sylvestres is a monotypic section containing only N. sylvestris and is c. 6.2 Ma from the plastid analysis and c. 7.7 Ma from the nuclear analysis. Tomentosae contains five species and the section yields age estimates of c. 13.0 Ma from the plastid analysis and c. 10.1 Ma from the nuclear analysis. Trigonophyllae contains two taxa (both subspecies of N. obtusifolia) with age estimates of c. 10.2 Ma from the plastid analysis and c. 10.1 Ma from the nuclear analysis. Undulatae contains a total of five species, and the section yields age estimates of c. 7.7 Ma from the plastid analysis. Data for this section are unavailable from the nuclear analysis. The difference between the age estimates yielded by the plastid and nuclear trees for these diploid sections is probably due to the alternative placement of the allopolyploids in the two analyses.
The results of geologically calibrated analyses are summarised in Online Resource 4—Geological Calibration Table. They show that the two geological calibration points yield allopolyploid ages that are generally slightly older than with the fossil-derived calibration points. Also, the error bars tend to be higher with the geological calibration particularly for the deeper nodes of the tree.
Discussion
Age estimates in Nicotiana
Early studies using protein data estimated the genus Nicotiana originated around 75–100 Ma (Uchiyama et al. 1977), but this now clearly seems to be an error and is totally refuted by published molecular clock studies (Särkinen et al. 2013; Wikström et al. 2001). Mummenhoff and Franzke (2007) used nrITS sequence divergence for N. section Suaveolentes (6.5% with a range of 0.2–15.0%) and applied a standard rate of 1% ITS divergence per 0.5–1.0 Myr. They concluded that these Australian Nicotiana species are only 3.25–6.50 Myr old. Ladiges et al. (2011) speculated that the age of Nicotiana sect. Suaveolentes was 15 Ma based on their current distribution in Australia and an assumption that their common ancestor was already broadly distributed across the continent before the development of the Eremaean Zone, which caused fragmentation and isolation of the resulting populations, leading to formation of the modern species in that region. This age assumption is highly inconsistent with Särkinen et al. (2013), and in fact 15 Ma is closer to the age of the Nicotiana–Symonanthus node. Our age estimates suggest that establishment of the progenitor of N. sect. Suaveolentes in Australia occurred near the end of a round of aridification, ca. 10–7 Mya (Byrne et al. 2008). This period was followed by a somewhat wetter period that lasted until 4 Mya, when a fully arid interior was established, and it is during this period that species diversification of N. sect. Suaveolentes occurred. This date is unusually late for speciation in the Eremaean Zone. Byrne et al. (2008) stated that there was no evidence to support increased rates of speciation during the Pleistocene (2.6 Mya until 12,000 years ago), but this is exactly when we detected an increased rate of speciation in N. sect. Suaveolentes. This hypothesis contrasts dramatically with that postulated by Ladiges et al. (2011), who assumed that the earlier molecular clock studies were incorrect, citing problems with accurate assignment of fossils as calibration points (Graur and Martin 2004; Heads 2005; Nelson and Ladiges 2009), but our study used two methods of calibration with two independent datasets that are largely congruent among themselves about the dates of origin/establishment of N. sect. Suaveolentes in Australia. This gives our estimates of ages more credence than other such molecular clock studies. The subsequent diversification of Suaveolentes represents a novel event: a group that has responded positively to the extreme drying of the Eremaean zone in the last two million years and diversified.
As noted above, the two datasets (plastid and nuclear) analysed in this study are roughly consistent and corroborate each other (see Table 1). These dates agree well with the volcanic island calibration points used in previous studies (Clarkson et al. 2005), which are generally older, as would be expected when comparing minimum dates from fossils with maximum dates from geological events. However, as with all time-calibrated phylogenetic approaches, the ages yielded are approximate, and inferences should be made in the context of the confidence intervals. We used a secondary calibration point due to a lack of fossil evidence for the genus, and this also introduced a degree of error. Another limitation inherent to this study stems from focusing on a genus of around 75 species because of the limited number of data points available. This can be contrasted with the much larger, angiosperm-wide, phylogenetic datasets analysed elsewhere (e.g. Tank et al. 2015). Accepting these limitations, we feel that calibrating at a node positioned at the base of the genus rather than out at the tips as with the geological dates (see Clarkson et al. (2005) and Online Resource 4—Geological Calibration Table) is an intrinsically more robust approach due to the fact that the whole tree is no longer scaled according to a short branch. The timeline of ages yielded with these data reveals the single polyploid species, N. tabacum, N. rustica and N. arentsii; all have mean age estimates of less than 1 Ma. Nicotiana sect. Polydicliae (two species) is 1.5 Ma, N. sect. Repandae (four species) is 4 Ma, and N. sect. Suaveolentes (35 species) is 6 Ma (Fig. 3). The dates yielded for nodes (see Online Resource 2 and 3) are generally lower than with geological calibration (see Clarkson et al., 2005, and Online Resource 4), but because geological calibration yields maximum ages, the two approaches are consistent.
A more accurate timeframe for speciation and polyploid events established by these results could aid the interpretation of newer approaches being applied to Nicotiana allopolyploids such as transcriptomic studies. A current focus is being given to the expression patterns of both young allopolyploids such as N. tabacum (Bombarely et al. 2012) and older allopolyploids such as N. benthamiana of section Suaveolentes (Nakasugi et al. 2013, 2014). In the following section, we review aspects of diploidisation that have been documented for allopolyploids of different ages in Nicotiana.
Characteristics of allopolyploids formed less than 1 Ma: Nicotiana tabacum, N. arentsii and N. rustica
These three recently formed allopolyploids and in particular N. tabacum have been extensively studied, and since they formed under 1 Ma, they represent an early stage of the diploidisation processes. When comparing N. tabacum, N. arentsii and N rustica to their progenitor diploids, these allopolyploid genomes are clearly additive. The two parental genomes can be completely differentiated using genomic in situ hybridisation (GISH) because their repetitive elements are still sufficiently similar to those in their progenitors (Lim et al. 2004). These three allopolyploids also show additivity, with respect to their progenitors in the 5S and 35S rDNA loci (Lim et al. 2004, 2007). Inter-genomic translocations have been detected in N. tabacum (Kovarik et al. 2012; Lim et al. 2004) but not in N. rustica and N. arentsii. Nicotiana tabacum may have undergone inter-genomic translocations because it has the most divergent diploid progenitors of these three allotetraploids, and therefore, translocations could form part of the post-WGD stabilisation process (Song et al. 1995).
Concerted evolution of rDNA has been documented in N. tabacum, N. arentsii and N. rustica (summarised in Kovarik et al. (2004)) resulting in the rDNA loci being overwritten in a few generations by one dominant progenitor copy. This is usually the maternal copy, but there is at least one example of the paternal copy dominating, in N. tabacum (Chase et al. 2003). The process of rDNA concerted evolution has been documented to occur rapidly after polypoid formation, within just four generations in synthetic tobacco lines (Kovarik et al. 2012; Skalicka et al. 2003). Loss of paternally derived genomic repeats was first identified in N. tabacum by Skalicka et al. (2005), and a subsequent study, using high-throughput sequencing methods, showed this paternal loss is widespread for most repeat types (Renny-Byfield et al. 2011). In N. rustica, repetitive DNA revealed evidence of reduction and rearrangement of repeats from one of the progenitor subgenomes (Lim et al. 2005). Transcriptomic data from N. tabacum have shown that there is little evidence for positive selection on homeologs, and therefore, neofunctionalisation of genes seems unlikely due to the young age of this allotetraploid. However, gene silencing of some homeologs was evident, suggesting some degree of modification to gene expression in this young allotetraploid (Bombarely et al. 2012). Genome size values (1C-values) show that the three polyploids are similar to the sum of their progenitor genomes, but they have experienced small amounts of downsizing in the range of 2–5% below additivity (Leitch et al. 2008).
Characteristics of Nicotiana section Polydicliae (c. 1.5 Ma)
Nicotiana sect. Polydicliae consists of two species, N. quadrivalvis and N. clevelandii, that are native to the deserts of western North America, and their range extends as far north as northern California (Goodspeed 1954). Diploidisation has not been extensively studied in this young pair of closely related allopolyploids. The mixing of repetitive elements of different progenitor origin on chromosomes (genomic homogenisation) has been observed using GISH (Lim et al. 2007). This homogenisation seen in sect. Polydicliae can be contrasted with the additivity observed in younger polyploids within the genus (see above) as here the two clear progenitor subgenomes cannot be fully discriminated. The subtelomeric chromatin of N. quadrivalvis only labels with its maternal progenitor type, indicating that these repetitive elements have replaced repeats of paternal origin (Lim et al. 2007). Genome size measurements indicate these allopolyploids are approximately the sum of their progenitors but with a small degree of increase (3–8%) above expectation (Leitch et al. 2008).
Characteristics of Nicotiana sect. Repandae (c. 4 Ma)
Overall genome size (1C-value) shifts were reported in N. sect. Repandae. These allotetraploid species are not the sum of their progenitor diploid genomes and have undergone downsizing in one species (N. nudicaulis) and unexpectedly significant upsizing (19–29%) in the remaining species with respect to their progenitors (Leitch et al. 2008). The elimination of low-copy-number repeats is ubiquitous in N. sect. Repandae, but changes in the high-copy-number repeats in particular have been linked with changes in overall genome size that account for the differences observed between the N. sect. Repandae species (Renny-Byfield et al. 2013). The differences in repeat abundance between species of section N. sect. Repandae also reflect phylogenetic distance (Dodsworth et al. 2016b, this issue).
Large numbers of novel transposable elements (TEs) were found to be common in all species of Repandae, but are absent from their progenitor diploid genomes. Therefore, the TEs probably originated in the early evolutionary stages of the polyploid stock before speciation into the four extant species and may be linked to early diploidisation (Parisod et al. 2012). The loss of entire rDNA loci has been documented in Repandae and also the mixing of repeats of different parental origin on chromosomes (Clarkson et al. 2005), but progenitor subgenomes can still be distinguished (Dodsworth et al. 2016b).
Characteristics of N. sect. Suaveolentes (c. 6 Ma)
This is the oldest and most diploidised section, which has diversified to create ~35 extant species. The section ranges from n = 15–24, which we interpret to represent the result of complex chromosome fusions and translocations, whereas (Goodspeed 1954) believed this range to at least partially be the result of several independent allotetraploid hybridisation events between progenitor species with different chromosome numbers. All phylogenetic results thus far demonstrate that this is not the case, and the formation of N. sect. Suaveolentes was a single event followed by subsequent speciation at the polyploid level. All species in the section are found in Australia and surrounding Pacific Islands with the exception of N. africana. This species is endemic to Namibia and is the earliest diverging lineage within the section based on all phylogenetic data.
The progenitor diploids of N. sect. Suaveolentes are difficult to determine partly because of the antiquity of this event. An ancestor of N. sylvestris has been identified as the paternal progenitor (Clarkson et al. 2010), but evidence based on gene trees suggests various conflicting scenarios for the maternal progenitor. The most likely interpretation of the data suggests that the maternal progenitor was a hybrid species formed from the ancestral species of the extant sections Noctiflorae and Petunioides (Kelly et al. 2013), although this does not fully account for the variation in all gene trees. It has been suggested that introgression between Suaveolentes species may be widespread (Marks et al. 2011b), making species delimitation and phylogeny reconstruction complex because these allopolyploids are hybridising at the polyploid level. However, in four years of fieldwork in Australia (South Australia, Western Australia and the Northern Territory), MWC and SD have yet to observe a specimen that would qualify as a hybrid. Each species appears morphologically and ecologically distinct, and speciation is likely to be ongoing, in contrast to the scenario described by Ladiges et al. (2011).
Biogeographic and distributional studies focusing on the N. sect. Suaveolentes native to Australia show that the species in the wet tropical north and down the eastern seaboard tend to have higher chromosome numbers (the ancestral state), whereas lower numbers predominate in the hot and arid centre of Australia. There is also evidence of genome size reduction from an ancestral 1C = 5 pg down to 1C = ~3 pg in many Australian taxa (Dodsworth, personal communication). Two separate draft genomes have been assembled for N. benthamiana and are available at www.solgenomics.net/ and www.benthgenome.qut.edu.au/. Transcriptomic data are available from ten tissue types in Nicotiana benthamiana (Nakasugi et al. 2013 2014). We are now at a stage where evidence from genomic and transcriptomic data can be combined to address broader questions of genomic evolution. However, the large-scale sequence datasets available (both transcriptomes and genomes) need to be combined together in a coherent framework addressing the questions arising from the effects of WGD, diploidisation and their relationship with polyploid age.
Concluding remarks
The genus Nicotiana provides a unique opportunity to study polyploids of different ages ranging from neopolyploids to paleopolyploids. Our current study provides a timeframe for all speciation events including estimating polyploid ages based on genetic distances from their diploid progenitors. These results suggest Nicotiana arentsii is the youngest polyploid at 0.4 Ma and the oldest polyploids are 6 Ma (N. sect. Suaveolentes), a date that agrees with previous molecular estimates. A progression of diploidisation events has been established for allopolyploids of different ages in Nicotiana (discussed in detail above), which stabilises and transforms their genomes (see Fig. 1 of Wendel 2015). Older polyploids in Nicotiana have undergone more speciation than younger ones (Fig. 3), and although with increasing time we might expect any lineage to steadily diversify, this curve appears steep (in particular due to the N. sect. Suaveolentes data point). The progenitors of N. sect. Suaveolentes (the oldest polyploids estimated at 6 Ma) are found in South America, and these polyploids have colonised new niches in Australia and some Pacific islands. In the second oldest group, N. sect. Repandae (estimated at 4.3 Ma), species have colonised new environments (e.g. the Revillagigedo Islands) from mainland Mexican diploid progenitors. We hypothesise that it is the processes associated with diploidisation occurring during the post-WGD period that led to an increase in the diversification rates millions of years after the WGD event (Dodsworth et al. 2016a). This phenomenon is analogous to the lag period between WGD and diversification described at higher taxonomic levels in angiosperms (Schranz et al. 2012; Soltis et al. 2009; Tank et al. 2015). However, it is still unclear whether it is solely diploidisation or this plus colonisation that has played the most important role in increasing diversification rates in N. sect. Suaveolentes.
Diploidisation progresses through a series of stages (described above) that are evident in the genus due to the natural range of allopolyploid ages documented. Only in the range of approximately 4–6 million years do the genomic benefits of polyploidy appear to be having a marked effect on speciation (Fig. 3). It remains unclear exactly what has enabled the progenitor of N. sect. Suaveolentes to radiate into 35 species, but the answer might lie in the documented chromosome number reductions that could be a response to harsh environments and high selection pressure. These rearrangements may have resulted in the formation of beneficial new linkage groups on recently fused chromosome arms (Stebbins 1950). Another explanation might be related to the colonisation of new relatively underexploited areas due to the progenitors of N. sect. Suaveolentes inhabiting South America; their subsequent colonisation of Australia, Pacific islands and Namibia opened up new terrain to them. Therefore, the availability of new niches is likely another factor related to their evolutionary success. One puzzle that still remains is why the earliest diverging lineage in section Suaveolentes, N. africana, found in Namibia, has not undergone a similar burst of speciation as its sister group in Australia. To our knowledge, this study is the first report of a substantial lag phase being investigated at the species level within a genus.
References
Bombarely A, Edwards KD, Sanchez-Tamburrino J, Mueller LA (2012) Deciphering the complex leaf transcriptome of the allotetraploid species Nicotiana tabacum: a phylogenomic perspective. BMC Genomics 13:406. doi:10.1186/1471-2164-13-406
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D et al (2014) BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Computat Biol 10:e1003537. doi:10.1371/journal.pcbi.1003537
Byrne M et al (2008) Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Molec Ecol 17:4398–4417
Chase MW et al (2003) Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot (Oxford) 92:107–127
Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW (2004) Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Molec Phylogen Evol 33:75–90
Clarkson JJ, Lim KY, Kovarik A, Knapp S, Chase M, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252
Clarkson JJ, Kelly LJ, Leitch AR, Knapp S, Chase MW (2010) Nuclear glutamine synthetase evolution in Nicotiana: phylogenetics and the origins of allotetraploid and homoploid (diploid) hybrids. Molec Phylogen Evol 55:99–112
Dodsworth S et al (2015) Genomic repeat abundances contain phylogenetic signal. Syst Biol 64:112–126
Dodsworth S, Chase MW, Leitch AR (2016a) Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot J Linn Soc 180:1–5
Dodsworth S, Jang T-S, Struebig M, Chase MW, Weiss-Schneeweiss H, Leitch AR (2016b) Genome-wide repeat dynamics reflect phylogenetic distance in closely related allotetraploid Nicotiana (Solanaceae). Pl Syst Evol (First Online). doi:10.1007/s00606-016-1356-9
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Goodspeed TH (1954) The genus Nicotiana. Chronica Botanica Company, Waltham
Graur D, Martin W (2004) Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet 20:80–86
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer Publishing, Sunderland, pp 43–63
Heads M (2005) Dating nodes on molecular phylogenies: a critique of molecular biogeography. Cladistics 21:62–78
Kelly LJ, Leitch AR, Clarkson JJ, Hunter RB, Knapp S, Chase MW (2010) Intergenic recombination events and evidence for hybrid speciation in Nicotiana (Solanaceae). Molec Biol Evol 27:781–799
Kelly LJ, Leitch AR, Clarkson JJ, Knapp S, Chase MW (2013) Reconstructing the complex evolutionary origin of wild allopolyploid tobaccos (Nicotiana section Suaveolentes). Evolution 67:80–94
Knapp S, Chase MW, Clarkson JJ (2004) Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae). Taxon 53:73–82
Kovarik A et al (2004) Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol J Linn Soc 82:615–625
Kovarik A, Renny-Byfield S, Grandbastien MA, Leitch AR (2012) Evolutionary implications of genome and karyotype restructuring in Nicotiana tabacum L. In: Soltis PS, Soltis DE (eds) Polyploidy and Genome Evolution. Springer, Berlin, pp 209–224
Ladiges PY, Marks CE, Nelson G (2011) Biogeography of Nicotiana section Suaveolentes (Solanaceae) reveals geographical tracks in arid Australia. J Biogeog 38:2066–2077
Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR (2008) The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot (Oxford) 101:805–814
Lim KY, Matyasek R, Kovarik A, Leitch AR (2004) Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc 82:599–606
Lim KY, Matyasek R, Kovarik A, Fulnecek J, Leitch AR (2005) Molecular cytogenetics and tandem repeat sequence evolution in the allopolyploid Nicotiana rustica compared with diploid progenitors N. paniculata and N. undulata. Cytogenet Genome Res 109:298–309
Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, Grandbastien MA, Leitch AR (2007) Sequence of events leading to near complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol 175:756–763
Marks CE (2010) Definition of South Pacific taxa of Nicotiana section Suaveolentes (Solanaceae). Muelleria 28:74–84
Marks CE, Ladiges PY, Newbigin E (2011a) Karyotypic variation in Nicotiana section Suaveolentes. Genet Resources Crop Evol 58:797–803. doi:10.1007/s10722-011-9724-3
Marks CE, Newbigin E, Ladiges PY (2011b) Comparative morphology and phylogeny of Nicotiana section Suaveolentes (Solanaceae) in Australia and the South Pacific. Austral Syst Bot 24:61–86
McCarthy EW et al (2015) The effect of polyploidy and hybridization on the evolution of floral colour in Nicotiana (Solanaceae). Ann Bot (Oxford) 115:1117–1131
McCarthy EW, Chase MW, Knapp S, Litt A, Leitch AR, Le Comber SC (2016) Transgressive phenotypes and generalist pollination in the floral evolution of Nicotiana polyploids. Nat Pl 2:16119. doi:10.1038/nplants.2016.119
Mummenhoff K, Franzke A (2007) Gone with the bird: late Tertiary and Quaternary intercontinental long-distance dispersal and allopolyploidization in plants. Syst Biodivers 5:255–260
Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE 8:e59534. doi:10.1371/journal.pone.0059534
Nakasugi K, Crowhurst R, Bally J, Waterhouse P (2014) Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS ONE 9:e91776. doi:10.1371/journal.pone.0091776
Nelson G, Ladiges PY (2009) Biogeography and the molecular dating game: a futile revival of phenetics? Bull Soc Géol France 180:39–43
Nylander JAA (2008) MrModeltest v2.3. Available at: https://github.com/nylander/MrModeltest2
Parisod C, Mhiri C, Lim KY, Clarkson JJ, Chase MW, Leitch AR, Grandbastien MA (2012) Differential dynamics of transposable elements during long-term diploidization of Nicotiana section Repandae (Solanaceae) allopolyploid genomes. PLoS ONE 7:e50352. doi:10.1371/journal.pone.0050352
Rambaut A (2014) FigTree v1.4.2. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Rambaut A, Drummond AJ (2014) Tree Annotator v1.8.2. Available at: http://beast.bio.ed.ac.uk/TreeAnnotator
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer v1.6. Available at: http://beast.bio.ed.ac.uk/Tracer
Renny-Byfield S et al (2011) Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Molec Biol Evol 28:2843–2854
Renny-Byfield S et al (2013) Diploidization and genome size change in allopolyploids is associated with differential dynamics of low and high-copy sequences. Pl J 74:829–839. doi:10.1111/tpj.12168
Särkinen T, Bohs L, Olmstead RG, Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol Biol 13:214. doi:10.1186/1471-2148-13-214
Schranz ME, Mohammadin S, Edger PP (2012) Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr Opin Pl Biol 15:147–153. doi:10.1016/j.pbi.2012.03.011
Skalicka K, Lim KY, Matyasek R, Koukalova B, Leitch AR, Kovarik A (2003) Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. Amer J Bot 90:988–996. doi:10.3732/ajb.90.7.988
Skalicka K, Lim KY, Matyasek R, Matzke AJM, Leitch AR, Kovarik A (2005) Preferential elimination of repeated DNA sequences from the paternal, N. tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol 166:291–303
Soltis DE, Soltis PS (1990) Isozymes in plant biology. Chapman and Hall, London
Soltis DE et al (2009) Polyploidy and angiosperm diversification. Amer J Bot 96:336–348. doi:10.3732/ajb.0800079
Soltis DE, Visger CJ, Marchant DB, Soltis PS (2016) Polyploidy: pitfalls and paths to a paradigm. Amer J Bot 103:1–21. doi:10.3732/ajb.1500501
Song KM, Lu P, Tang KL, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Stebbins GL (1950) Variation and evolution in plants. Columbia University Press, New York
Swofford DL (2001) PAUP* 4.0: Phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland
Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, Brown JW (2015) Nested radiations and the pulse of angiosperm diversification: increased diversification rates follow whole genome duplications. New Phytol 207:454–467. doi:10.1111/nph.13491
Uchiyama H, Chen K, Wildman SG (1977) Polypeptide composition of fraction I protein as an aid in the study of plant evolution. Stadler Symposium 9:83–99
Wendel JF (2015) The wondrous cycles of polyploidy in plants. Amer J Bot 102:1753–1756. doi:10.3732/ajb.1500320
Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc Roy Soc Biol Sci Ser B 268:2211–2220. doi:10.1098/rspb.2001.1782
Acknowledgements
We would like to thank the Bentham-Moxon Trust for fully funding the costs for JC to attend the International Polyploidy and Hybridisation meeting in 2016. Thanks to Andrew Leitch for useful discussions on diploidisation in Nicotiana and Laura Kelly for sharing LFY sequence data. We are grateful that the CIPRES Science Gateway provided us with server access for our phylogenetic analyses. Thanks also to Dion Devey, Robyn Cowan and Laura Martinez-suz for providing technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that we have no conflict of interest.
Human and animal rights
This article did not involve any testing on humans or animals.
Additional information
Handling editor: Ales Kovarik.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Figure showing combined nuclear (GS and LFY) parsimony bootstrap tree. Elevated support is used as an indication of congruence between the two datasets.
Online Resource 2. Time-calibrated plastid tree displaying mean age estimates for all nodes. Yielded from Bayesian relaxed molecular clock method implemented in BEAST2 (Bouckaert et al. 2014) and using fossil derived calibration. Tree topology identical to Fig. 1 but here node ages are displayed rather than HPD values.
Online Resource 3. Time-calibrated nuclear tree displaying mean age estimates for all nodes. Yielded from Bayesian relaxed molecular clock method implemented in BEAST2 (Bouckaert et al. 2014) and using fossil derived calibration. Tree topology identical to Fig. 2 but here node ages are displayed rather than HPD values.
Online Resource 4. Table showing node ages for allopolyploids yielded by time calibrated phylogenetic analyses (BEAST2) using geological calibration points. The node associated with N. cordifolia–N. solanifolia was calibrated at 2.4 million years and the N. nesophila, N. stocktonii–N. repanda node was calibrated at 1.1 million years.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Clarkson, J.J., Dodsworth, S. & Chase, M.W. Time-calibrated phylogenetic trees establish a lag between polyploidisation and diversification in Nicotiana (Solanaceae). Plant Syst Evol 303, 1001–1012 (2017). https://doi.org/10.1007/s00606-017-1416-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1416-9