Abstract
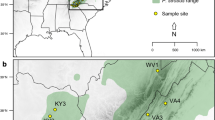
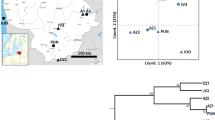
Twenty-one populations (555 individuals) covering the entire native range of Pinus mugo Turra (dwarf mountain pine) were investigated for genetic variation scored at 13 nuclear microsatellite markers (nSSRs). The main objective of the present study was to determine the genetic structure across the present distribution of the species and locate populations of different genetic compositions. Most of the genetic variation was observed within the populations (95%). The assignment of populations based on Bayesian clustering methods revealed that the Sudeten populations of P. mugo form a separate genetic cluster. These stands have likely been established through the founder effects of Alpine migrants. The distribution and level of SSR polymorphisms, along with no evidence of isolation by distance or phylogeographic structure, indicate that the present populations of P. mugo have diverged relatively recently and originate from a larger glacial distribution of the species. One peripheral stand from Italy had the lowest values of most calculated genetic variation indices. This stand could therefore be more susceptible to genetic drift and a negative impact of predicted environmental changes. We discuss our findings with respect to previously published results on the genetic and morphological variation of P. mugo and with consideration for the conservation genetics of the species.

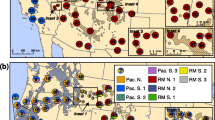
(modified after Critchfield and Little (1971))

Similar content being viewed by others
References
Afzal-Rafii Z, Dodd RS (2007) Chloroplast DNA supports a hypothesis of glacial refugia over postglacial recolonization in disjunct populations of black pine (Pinus nigra) in Western Europe. Molec Ecol 16:723–736. doi:10.1111/j.1365-294X.2006.03183.x
Boratyńska K, Boratyński A (2007) Taxonomic differences among closely related pines Pinus sylvestris, P. mugo, P. uncinata, P. rotundata and P. uliginosa as revealed in needle sclerenchyma cells. Flora 202:555–569. doi:10.1016/j.flora.2006.11.004
Boratyńska K, Muchewicz E, Drojma M (2004) Pinus mugo Turra geographic differentiation based on needle characters. Dendrobiology 51:9–17
Boratyńska K, Dzialuk A, Lewandowski A, Marcysiak K, Jasińska AK, Sobierajska K, Tomaszewski D, Burczyk J, Boratyński A (2014) Geographic distribution of quantitative traits variation and genetic variability in natural populations of Pinus mugo in Central Europe. Dendrobiology 72:65–84. doi:10.12657/denbio.072.006
Boratyńska K, Jasińska AK, Boratyński A (2015) Taxonomic and geographic differentiation of Pinus mugo complex on the needle characteristics. Syst Biodivers 13:581–595. doi:10.1080/14772000.2015.1058300
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Molec Biol Evol 24:621–631. doi:10.1093/molbev/msl191
Chapuis MP, Lecoq M, Michalakis Y, Loiseau AG, Sword G, Piry S (2008) Do outbreaks affect genetic population structure? A worldwide survey in Locusta migratoria, a pest plagued by microsatellite null alleles. Molec Ecol 17:3640–3653. doi:10.1111/j.1365-294X.2008.03869.x
Chybicki I (2015) INEST 2.0. Available at: http://www.ukw.edu.pl/pracownicy/strona/igor_chybicki/software_ukw/
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Critchfield WB, Little EL (1971) Geographic distribution of the pines of the world. U.S. Department of Agriculture, Washington
Di Rienzo AA, Peterson C, Garza JC, Valdes AM, Slatkin M, Freime RB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256. doi:10.1007/BF00220937
Dzialuk A, Boratyński A, Boratyńska K, Burczyk J (2012) Geographic patterns of genetic diversity of Pinus mugo (Pinaceae) in Central European mountains. Dendrobiology 68:31–41
Earl DA, von Holdt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resources 4:359–361. doi:10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molec Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
García-Amorena I, Gómez Manzaneque F, Rubiales JM, Granja HM, Soares de Carvalho G, Morla C (2007) The Late Quaternary coastal forests of western Iberia: a study of their macroremains. Palaeogeogr Palaeoclimatol Palaeoecol 254:448–461. doi:10.1016/j.palaeo.2007.07.003
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at: http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from Goudet (1995)
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467. doi:10.1111/j.1461-0248.2005.00739.x
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans Ser B Biol Sci 351:1291–1298
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molec Ecol Notes 2:618–620. doi:10.1046/j.1471-8286.2002.00305.x
Hardy OJ, Charbonnel N, Fréville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Heuertz M, Teufel J, González-Martínez SC, Soto A, Fady B, Alía R, Vendramin GG (2010) Geography determines genetic relationships between species of mountain pine (Pinus mugo complex) in western Europe. J Biogeogr 37:541–556. doi:10.1111/j.1365-2699.2009.02223.x
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Latałowa M, Tobolski K, Nalepka D (2004) Pinus L. subgenus Pinus (subgen. Diploxylon (Koehne) Pilger)—Pine. In: Ralska-Jasiewiczowa M (ed) Late glacial and Holocene history of vegetation in Poland based on isopollen maps. W. Szafer Institute of Botany, Kraków, pp 165–177
Lewandowski A, Boratyński A, Mejnartowicz L (2000) Allozyme investigations on the genetic differentiation between closely related pines—Pinus sylvestris, P. mugo, P. uncinata, and P. uliginosa (Pinaceae). Pl Syst Evol 221:15–24. doi:10.1007/BF01086377
Lowry DB (2010) Landscape evolutionary genomics. Biol Lett 6:502–504. doi:10.1098/rsbl.2009.0969
Luikart G, Cornuet J-M (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biol 12:228–237
Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ Jr, Roth A, Bagley MJ, Nacci DE (2010) Population genetic diversity and fitness in multiple environments. BMC Evol Biol 10:205. doi:10.1186/1471-2148-10-205
Matschiner M, Salzburger W (2009) TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25:1982–1983. doi:10.1093/bioinformatics/btp303
Monteleone I, Ferrazzini D, Belletti P (2006) Effectiveness of neutral RAPD markers to detect genetic divergence between the subspecies uncinata and mugo of Pinus mugo Turra. Silva Fenn 40:391–406
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122. doi:10.1038/nrg2931
Nosil P, Feder JL (2013) Genome evolution and speciation: toward quantitative descriptions of pattern and process. Evolution 67:2461–2467. doi:10.1111/evo.12191
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Molec Ecol 22:5983–5999. doi:10.1111/mec.12561
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molec Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carrillo C, Pauli JN, Palsbøll PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Molec Ecol 21:3403–3418. doi:10.1111/j.1365-294X.2012.05635.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Robledo-Arnuncio JJ, Collada C, Alía R, Gil L (2005) Genetic structure of montane isolates of Pinus sylvestris L. in a Mediterranean refugial area. J Biogeogr 32:595–605. doi:10.1111/j.1365-2699.2004.01196.x
Rousset F (2008) Genepop’007: a complete re-implementation of the Genepop software for Windows and Linux. Molec Ecol Resources 8:103–106. doi:10.1111/j.1471-8286.2007.01931.x
Sannikov SN, Petrova IV, Schweingruber F, Egorov EV, Parpan TV (2011) Genetic differentiation of Pinus mugo Turra and P. sylvestris L. populations in the Ukrainian Carpathians and the Swiss Alps. Russ J Ecol 42:270–276. doi:10.1134/s1067413611040151
Schoville SD, Bonin A, François O, Lobreaux S, Melodelima C, Manel S (2012) Adaptive genetic variation on the landscape: methods and cases. Annual Rev Ecol Evol Syst 43:23–43. doi:10.1146/annurev-ecolsys-110411-160248
Sinclair WT, Morman JD, Ennos RA (1998) Multiple origins for Scots pine (Pinus sylvestris L.) in Scotland evidence from mitochondrial DNA variation. Heredity 80:233–240. doi:10.1046/j.1365-2540.1998.00287.x
Slavov GT, Zhelev P (2004) Allozyme variation, differentiation, and inbreeding in populations of Pinus mugo in Bulgaria. Canad J Forest Res 34:2611–2617. doi:10.1139/x04-127
Sobierajska K, Boratyńska K (2008) Variability of needle characteristics of Pinus mugo Turra populations in the Karkonosze Mountains in Poland. Dendrobiology 59:41–49
Sobierajska K, Boratyńska K, Marcysiak K (2010) Variation of cone characters in Pinus mugo (Pinaceae) populations in the Giant Mountains (Karkonosze, Sudetes). Dendrobiology 63:33–41
Soranzo N, Alía R, Provan J, Powell W (2000) Patterns of variation at a mitochondrial sequence-tagged-site locus provides new insights into the postglacial history of European Pinus sylvestris populations. Molec Ecol 9:1205–1211. doi:10.1046/j.1365-294x.2000.00994.x
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molec Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Vendramin GG, Degen B, Petit RJ, Anzidei M, Madaghiele A, Ziegenhagen B (1999) High level of variation at Abies alba chloroplast microsatellite loci in Europe. Molec Ecol 8:1117–1126. doi:10.1046/j.1365-294x.1999.00666.x
Wachowiak W, Boratyńska K, Cavers S (2013) Geographical patterns of nucleotide diversity and population differentiation in three closely related European pine species in the Pinus mugo complex. Bot J Linn Soc 172:225–238. doi:10.1111/boj.12049
Willis KJ, Bennett KD, Birks HJB (1998) The late Quaternary dynamics of pines in Europe. In: Richardson DM (ed) Ecology and biogeography of Pinus. Cambridge University Press, Cambridge, pp 107–121
Willis KJ, Rudner E, Sümegi P (2000) The full-glacial forests of Central and Southeastern Europe. Quatern Res 53:203–213. doi:10.1006/qres.1999.2119
Żukowska WB, Wachowiak W (2016) Utility of closely related taxa for genetic studies of adaptive variation and speciation: current state and perspectives in plants with focus on forest tree species. J Syst Evol 54:17–28
Acknowledgements
The authors would like to thank K. Boratyńska and A. Boratyński for the collection of the plant material and M. Litkowiec for technical support and valuable comments concerning the data analysis. The work was financially supported through a grant from the Polish National Science Centre (Grant No. DEC-2012/05/E/NZ9/03476).
Funding
This study was funded through a grant from the Polish National Science Centre (Grant No. DEC-2012/05/E/NZ9/03476).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Handling editor: Andreas Tribsch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Characteristics of the 22 nSSR markers initially tested.
Online Resource 2. Private alleles detected in the analysed Pinus mugo populations.
Online Resource 3. Pairwise matrix of F ST Excluding Null Alleles (F STnull) among the Pinus mugo populations.
Rights and permissions
About this article
Cite this article
Żukowska, W.B., Wachowiak, W. Nuclear microsatellite markers reveal the low genetic structure of Pinus mugo Turra (dwarf mountain pine) populations in Europe. Plant Syst Evol 303, 641–651 (2017). https://doi.org/10.1007/s00606-017-1395-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1395-x