Abstract
Polystachya section Superpositae is a group of montane and submontane African epiphytes with thirteen species (including the newly described in this paper). During a scientific expedition to Bamenda Highland, Cameroon a new species, Polystachya bamendae, was found and it is described here. The new entity was compared with 398 herbarium specimens representing the section, including the type collections. The molecular analyses of the ITS and matK strongly support the results of morphological studies and reveal close relationship between the new species and the species of the section Superpositae. Two hypotheses supporting function of the matK gene in the examined species are presented. The phylogenetic and morphological relation of Polystachya bamendae to P. lindblomii and P. superposita is discussed. The distribution range of the section is analyzed and a discussion about the distribution of the species is presented. A distribution map of the section and dichotomous key to the species of Superpositae are provided. The occurrence of the species of the section in two Biodiversity Hotspots (Guinea Forests of West Africa and Eastern Afromontane) is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Polystachya Hook. includes 233 species worldwide, of which nearly 90 % occurs in Africa (Mytnik-Ejsmont 2011). Representatives of the genus are sympodial, epiphytic, lithophytic or occasionally terrestrial herbs with terminal inflorescences growing always out of the top of the pseudobulb, with non-resupinate flowers. The flowers, predominantly small, are characterized by a mentum, which is a fusion of lateral sepals and the column foot, and three-lobed lip with usually oblong callus.
The section Superpositae Kraenzl. is one of 13 sections of Polystachya (Mytnik-Ejsmont 2011). It was described by Kraenzlin in the first monographic treatment of the genus (1926). The representatives of the section are montane and submontane epiphytes characterized i.e., by unique vegetative characters—fusiform pseudobulbs united in an erect or pendulous sympodium, each succeeding one arising from the middle or apical part of the proceeding one, 2–8-leaved, short unbranched or paniculate inflorescence, free part of gynostemium 1–2.5(5) mm high, and prominent column foot 4–5 mm long.
The section is divided into three subsections (Mytnik-Ejsmont 2011). The nominal subsection includes five species, P. eurygnatha Summerh., P. fusiformis (Thouars) Lindl., P. simplex Rendle, P. stewartiana Geerinck, and P. superposita Rchb.f. They are plants with pseudobulbs 3 mm in diameter at least, narrowly elliptic leaves, paniculate, 20–80-flowered inflorescence, lip shortly and broadly clawed, most often without any callus. Within the subsection Spatellae Mytnik there is classified seven species, P. spatella Kraenzl., P. ruwenzoriensis Rendle, P. lindblomii Schltr., P. pudorina P.J.Cribb, P. tridentata Summerh., P. aethiopica P.J.Cribb and newly described P. bamendae Szlachetko, Baranow & Mytnik. They can be defined by possession of slender stems up to 3 mm in diameter, scarcely thickened, narrowly oblong ligulate to linear leaves, unbranched inflorescence 1–16-flowered (most often consisting of 3–10 flowers), and long-clawed lip furnished with a fleshy callus. The subsection Kermesinae Mytnik is a monotypic taxon represented by P. kermesina Kraenzl., one of the showiest species within the subtribe. The species is atypical for Superpositae for its flower color and lip structure, however, due to its superposed, slender, 2–3-leaved pseudobulbs, linear leaves, short 3–5-flowered inflorescence, gynostemium structure and distribution range as well, Mytnik-Ejsmont (2011) decided to keep P. kermesina in Superpositae.
The section was revised once by Cribb (1979). The author presented a synopsis of the group and proposed three informal subgroups within the section. He included six species to the first group (P. spatella, P. ruwenzoriensis, P. lindblomii, P. pudorina, P. tridentata, and P. aethiopica), they were characterized by slender cylindrical stems scarcely pseudobulbous, narrow, grass-like leaves, medium-sized flowers, unbranched inflorescence, and the long-clawed lip furnished with a fleshy simple or lobed porrect callus. The plants with stouter pseudobulbs, broader, narrowly elliptic leaves, a paniculate inflorescence, and small flowers with a shortly clawed lip with or without a small unlobed callus were included in the second group (P. simplex, P. eurygnatha, P. fusiformis, and P. superposita). The last monotypic group was represented by Malgasian P. oreocharis Kraenzl.
The latter species, however, was excluded from the section by Mytnik-Ejsmont (2011). It was classified within Superpositae by many authors (Kraenzlin 1926; Cribb 1979, 1984) though it is a completely odd taxon. This Malgasian endemic is characterized by non-pseudobulbous stems completely covered by the imbricating leaf bases, many distichously arranged leaves, geniculate lip with strongly undulate margins and horn-shaped basal callus.
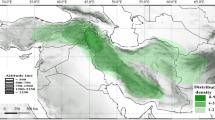
Apart from Polystachya superposita and P. bamendae, the novelty described in this paper, being endemic to Cameroon Highlands and P. fusiformis being widely distributed throughout tropical and subtropical Africa and Madagascar, vast majority of the species of Superpositae are confined to East African Rift System (Eastern Afromontane Biodiversity Hotspot), which is a center of high concentration of the species of the section. They occur mainly at higher elevations, most often from 850 to 3,200 m above sea level from Ethiopia to Mozambique. Five East African species of the section (P. eurugnatha, P. kermesina, P. ruwenzoriensis, P. stewartiana, and P. tridentata) are confined in their distribution to the Albertine Rift (=Western Rift Valley), the western branch of the East African Rift. The Albertine Rift, the region of a large number of endemic species, has been identified as an ecoregion by the World Wildlife Fund and a Biodiversity Hotspot by Conservation International (2005). Another representative of the section, Polystachya pudorina, is an endemic to part of the eastern branch of the East African Rift, the Usambara Mountains, which are unique ancient mountain region covered with the tropical forest. Due to a lack of glaciations and a relatively consistent climate, the rainforest of Usambaras has gone through a long-term evolution resulting in an impressive amount of endemism (Lovett 1993). Polystachya simplex and P. lindblomii, occur in both branches of the East African Rift along its whole length, from Ethiopian Highlands to Mozambique. Polystachya spatella, though known in both Western and Eastern Rift Valley, is confined in its distribution to the region of Victoria Lake. The species is one of the very few polystachyas growing above 3,000 m.a.s.l. It was even found above the evergreen forest belt, in the bamboo zone in Ruwenzori Mountains. The only species occurring exclusively in Ethiopia is Polystcahya aethiopica, being found in two ecoregions (WWF 2012): the Ethiopian montane forests (up to 1,800 m elevation) and the Ethiopian montane grasslands and woodlands (above 1,800 m).
Today the section includes thirteen species (including the new entity). The new species described here was found during a scientific expedition to Cameroon, in which Szlachetko and Baranow have been conducted taxonomic research of Orchidaceae. The country is among the top ten countries in Africa for biodiversity due to its montane forests, where a high number of endemic plant species occur (BirdLife International 2010) and to its strong latitudinal gradient of precipitation. Now the section Superpositae is represented by three species in Cameroon.
Material examined
Morphological study. The study presented here is based on the examination of the plant material collected in the field and 398 herbarium specimens from fifteen herbaria: B, BM, BR, BRLU, C, HBG, K, L, MA, MO, P, UGDA, W, WAG and YA (acronyms according to Thiers 2008, continuously updated). We observed some living plants in WAG and C. The specimen representing the new species was found in south-east of the Bamenda Highlands, about 2 h walking from the village Big Babanki to Czech Field Station.
DNA material. DNA sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Two data matrices, ITS and rps16-matK were taken from PopSet database no 298263940 (Russell et al. 2010a) and 270484601 (Russell et al. 2010b), respectively. The rps16-matK data matrix was trimmed for part of matK gene only and then analyzed. Four species (Adrorhizon purpurascens, Sirhookera lanceolata, Bromheadia finlaysoniana, and B. srilankensis) were selected as outgroup for matK and two species (Adrorhizon purpurascens and Sirhookera lanceolata) were selected as outgroup for ITS. In addition, three species (ITS) were taken from GenBank: Polystachya spatella HM018555, P. campyloglossa HM041027, P. lawrenceana HM018549. One species (ITS), P. fusiformis (id 068) was taken from data matrix from Anton Russell. Plant material for new species was collected during field expedition in 2011 to Cameroon. DNA from this specimen was extracted from silica dried leaves (Chase and Hillis 1991) using DNA easy Plant Mini Kit (A&A Biotechnology) following manufacturer protocol. ITS and part of matK gene (approximately 1,300 bp) were sequenced. Sequences were deposited in GenBank under accession number KC339535 (matK) and KC339536 (ITS).
Methods
DNA amplification and sequencing. Polymerase chain reaction (PCR) amplifications were carried out in a total volume of 25 μl containing 2.5 μl 10× buffer, 1 μl 25 mM MgCl2, 1 μl 5 mM dNTPs, 0.5 μl of 10 μM of each primers and 1 U of Red Perpetual DNA polymerase (Eurx, Poland) and H2O. The ITS region (ITS1-5.8S-ITS2) was amplified via PCR using the primers 101 AB and 102 AB (Douzery et al. 1999), the matK was amplified with the following two primers: 19F (Molvray et al. 2000) and 1,326R (Cuénoud et al. 2002). The PCR cycling conditions for ITS and matK were template premelting at 94 °C for 3 min followed by 30 cycles of denaturation at 95 °C for 45 s, primer annealing at 52 °C for 1 min, followed by primer extension at 72 °C for 90 s. Final extension step was of 7 min at 72 °C. Amplified products were cleaned with High Pure PCR Product Purification Kit (Roche Diagnostic GmbH, Mannheim, Germany) following manufacturer protocol. Cycle sequencing was carried out using Big Dye Terminator v 3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., ABI, Warrington, Cheshire, UK) with the same primers used for PCR amplification: 2.0 μl of 5× sequencing buffer, 1 μl of Big Dye terminator with 1.5 μl of 10 μM primer (1.5 pmol), 1–2 μl of amplified product (30–90 ng/μl), and H2O in a total of 10 μl reaction volume. Cycle sequencing conditions were as follows: 25 cycles each with 15 s denaturation (94 °C), 5 s annealing (50 °C), and 4 min elongation (60 °C). The sequences were generated on an ABI 3720 automated capillary DNA sequencer. Both strands were sequenced to assure accuracy in base calling. Sequence Navigator (ABI) was used to edit the sequences, and the two complementary strands were assembled using AutoAssembler (ABI).
Sequence alignment and phylogenetic analyses. The ITS and matK sequences were aligned by eye using Seaview (Galtier et al. 1996). Maximum parsimony analyses were undertaken on two matrices separately and combined analyses using heuristic searches in PAUP* version beta 10 (Swofford 2000) with tree-bisection-reconnection (TBR) branch swapping and the MULTREES (holding multiple trees) option in effect with 1,000 replicates of random sequence addition, but saving only 10 trees for each replicate to reduce the time spent in swapping large numbers of suboptimal trees. Standard tree parameters were estimated (tree length, consistency index, CI, and retention index, RI). Internal support of clades was evaluated using non-parametric bootstrapping (Felsenstein 1985) with 1,000 replicates and the same settings as above, except for simple sequence addition. All characters were treated as unordered and equally weighted (Fitch 1971).
Morphological study. Standard procedure of preparing the herbarium material to facilitate stereomicroscopic observation was applied. The following vegetative characters of individual plants were analyzed: stem (height, shape, presence of glandular hairs), leaves (number, size, shape), sheaths (number, shape, size), inflorescence (size, density), floral bracts (size, shape, presence of glandular hairs) flowers, taken from the middle part of the inflorescence (presence of glandular hairs, size of pedicel and ovary, height and shape of mentum (spur), size and shape of lateral sepals, dorsal sepal, petals, and lip), as well as gynostemium. Particular parts of the flower were boiled, dissected, measured and drawn under a stereomicroscope. The results were then analyzed and compared with the type material, diagnoses and original illustrations. The database of the drawings and photographs of all studied specimens is available in the first author’s archives. For all species type material was available.
Results
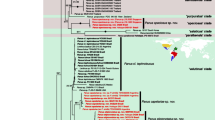
Statistics for ITS and matK and combined data matrices are separated by “/”. The number of analyzed taxa was 75/129/15, respectively. The aligned matrix comprised 860/1,326/2,158 characters of which 295/263/295 were variable and 195/162/120 were potentially parsimonial informative. The number of the most parsimonious trees were >10,000 for ITS and matK and 3 for combined analysis. Tree length was 702/414/360, consistency index (CI) = 0.6/0.7/0.7, and retention index (RI) = 0.8/0.9/0.8. One of the most parsimonious trees is depicted in Figs. 1, 2 and 3. Bootstrap support (BS) ≥50 % is given for supported clades below branches. Clades that collapse in the strict consensus tree are marked by an arrow. Both analyses indicate close relationships between new species and Polystachya section Superpositae. The bootstrap support for mentioned clade is low (BS <50/=50/56). Except P. lindblomii, all sequences from the Superpositae clade in matK gene possess 19 bp indel (duplication event). The indel occurrence causes a change of frameshift and as a natural consequence, an activation of the stop codon. The movement of the frameshift by one nucleotide downstream in the species possessing the indel enables an obtainment in silico the amino-acid sequence, which is homological to the sequences of the rest of the species and it does not have the stop codon within the gene sequence.
One of the most parsimonious trees in ITS analysis (L = 702, CI = 0.6 and RI = 0.8) for Polystachyinae. New species is highlighted in bold. The numbers above branches are Fitch branch lengths. Bootstrap percentages >50 are given for supported clades below branches. Arrows indicate clades that collapse in the strict consensus tree
One of the most parsimonious trees in matK analysis (L = 414, CI = 0.7 and RI = 0.9) for Polystachyinae. The new species is highlighted in bold. The numbers above branches are Fitch branch lengths. Bootstrap percentages >50 are given for supported clades below branches. Arrows indicate clades that collapse in the strict consensus tree
One of the most parsimonious trees in combined (matK + ITS) analysis (L = 360, CI = 0.8 and RI = 0.9) for Polystachyinae. The new species is highlighted in bold. The numbers above branches are Fitch branch lengths. Bootstrap percentages >50 are given for supported clades below branches—combined/ITS/matK, respectively. Arrows indicate clades that collapse in the strict consensus tree
Taxonomic treatment
Polystachya Hook. sect. Superpositae Kraenzl. subsect. Spatellae Mytnik
Polystachya bamendae Szlachetko, Baranow & Mytnik, sp. nov.
Type: Cameroon. North West Province: Bamenda Highlands about 2 h walking from the village Big Babanki to the Czech Field Station, in degraded secondary forest, ca 2–3 m above the ground on Eucalyptus trunk, elev. 2,150 m, 11–26 Nov 2011, fl. Sep–Oct 2012, Szlachetko & Baranow 9465 (holotype: UGDA; drawing of the holotype: BRLU, K, YA).
Polystachya bamendae is related to P. lindbomii, but the former species has the fractiflex rachis, prominent lip lateral lobes and the lip middle lobe 3 times wider than long.
Etymology: In reference to the place where the type specimen was collected.
En epiphytic plant up to 20 cm long. Pseudobulbs 7–11 cm long, 0.3–0.6 cm in diameter, fusiform, each succeeding one arising from the middle or apical part of the proceeding one. Leaves up to 6.5 cm long, 0.7 cm wide, narrowly lanceolate to linear-lanceolate, shortly acuminate. Inflorescence up to 8 cm long, branched, 4–20-flowered; rachis fractiflex, pubescent. Floral bracts 3 mm long, 4 mm wide, caudate. Flowers sparsely pubescent, fleshy, green with white apices of petals, lip yellow-white with yellow callus in the center. Pedicel and ovary 3–4 mm long, sparsely pubescent. Dorsal sepal 4 mm long, 2.8 mm wide, oblong-elliptic, caudate, apex 1.5 mm long. Lateral sepals 4.5–5 mm long, 2 mm wide, saccate, lance-ovate, caudate, apex 1–1.5 mm long. Mentum (spur) deeply bilobed, up to 4 mm long, 2 mm wide, rounded, saccate. Petals 3.5 mm long, 1 mm wide, narrowly elliptic, acute. Lip 4.2–6 mm long, 4–6 mm wide, 3-lobed, glabrous, strongly fleshy in the middle, squeezed in the middle on the outer surface, unguiculate, claw 2.5 mm long, 1.8 mm wide, ribbon-like, strongly fleshy along the midnerve; lateral lobes 2–2.5 mm long, 1.3–20 mm wide, widely obovate, obtuse, widely divergent; the middle lobe 2 mm long, 4.5–6 mm wide, transversely widely oblong, crenulate on margins, 4-lobed, emarginate with an apex 0.5–0.6 mm long on outer surface; the callus 1–2 mm long, 1 m high, fleshy bilobed ridge along the midnerve in the middle of the lip. Column foot 2–2.5 mm long, column 1.4–2 mm long, stout, anther 1.1 mm long and wide, beak-like, anther partitions prominent, lemon-shaped, 0.5 mm long, 0.3 mm wide; rostellum remnant retuse (Fig. 4).
Polystachya bamendae Szlachetko, Baranow & Mytnik a habit, b inflorescences and leaves, c leaf, d floral bract, e flower, f mentum, g lateral sepal, h dorsal sepal, i petal, j flower lacking lateral sepals, k lip, l trichomes on sepals, m rostellum remnant, n anther top view, o anther bottom view, p anther partition, r gynostemium
The morphological differences between the new species and the other taxa classified within the subsection Spatellae are presented in the Table 1.
Key to the species of Polystachya section Superpositae:
-
1.
Flowers orange to scarlet, fleshy, almost flat when opened, lip geniculate Polystachya kermesina
-
1.*
Flowers white, yellow, green, pink or purple, not widely opening, lip not geniculate 2
-
2.
Inflorescence paniculate, lip shortly clawed 3
-
2.*
Inflorescence simple, lip long-clawed 7
-
3.
Flowers fairly large, mentum 4 mm long or more, lip 15–18 mm long Polystachya stewartiana
-
3.*
Flowers small, mentum less than 4 mm long, lip 2–6 mm long 4
-
4.
Bracts 1–2.5 mm long, mentum less than 2 mm long, rounded 5
-
4.*
Bracts 3–7 mm long, mentum more than 2.5 mm long, conical 6
-
5.
Pseudobulbs 2–4-leaved, leaves linear-lanceolate, 4–9 cm long, lip with a callus Polystachya suerposita
-
5.*
Pseudobulbs 4–7-leaved, leaves oblong-lanceolate, 5–16 cm long, lip without a callus Polystachya fusiformis
-
6.
Sepals long acuminate, lip lateral lobes oblong, middle lobe acuminate Polystachya simplex
-
6.*
Sepals acute or subacute, lip lateral lobes obscure, middle lobe subacute or shortly mucronate Polystachya eurygnatha
-
7.
Ovary densely pubescent 8
-
7.*
Ovary glabrous or very sparsely pubescent 11
-
8.
Leaf more than 6.5 mm wide 9
-
8.*
Leaf less than 6 mm wide 10
-
9.
Lip callus 3-toothed, lip surface glabrous Polystachya tridentata
-
9.*
Lip callus simple, lip pubescent Polystachya aethiopica
-
10.
Flowers yellow-green, petals linear Polystachya lindblomii
-
10.*
Flowers pink or white with a crimson column, petals ovate-elliptic Polystachya pudorina
-
11.
Lip middle lobe markedly longer than wide Polystachya ruwenzoriensis
-
11.*
Lip middle lobe markedly wider than long 12
-
12.
Mentum more than 4.5 mm long, slightly bifid, lip middle lobe oblate Polystachya spatella
-
12.*
Mentum up to 4 mm long, deeply bilobed, lip middle lobe transversely widely oblong Polystachya bamendae
Polystachya bamendae, as well as P. superposita, are species endemic to submontane and montane forest of Cameroon (a part of Guinea Forests of West Africa Biodiversity Hotspot), (Figs. 5, 6). The representative of the newly described species was collected in the south-eastern part of the Bamenda Highlands, where the remaining patches of montane forest are very small (up to 500 ha, BirdLife International 2010), but biologically very important. As the narrowly endemic species, both Polystachya bamendae and P. superposita, should become a subject of very careful conservation action. Cameroonian Highland forests, where the species occur, are being cleared for firewood, timber and to create farmland. The Kilum-Ijim Forest, where two specimens of P. superposita were found, is the largest remaining patch of Afro-montane forest in West Africa being a very important area for biodiversity. Despite the same distribution range, both species are rather easy to distinguish morphologically. Polystachya bamendae is characterized by the fractiflex rachis, pubescent flowers, prominent deeply bilobed mentum up to 4 mm long and a fleshy clawed lip. The lip lateral lobes of the latter species are outspread, up to 2.5 mm long and 2 mm wide, the lip middle lobe is at least twice wider than long. The lip is squeezed in the middle and furnished with a prominent fleshy callus.
Discussion
Both plastid and nuclear ITS data indicate common origin of the species representing the section Superpositae. According to the results of the DNA analyses the most closely related species to Polystachya bamendae is P. lindblomii. The results of morphological analysis support the relation. Both species are erect or pendulous epiphytic herbs forming clumps with linear to linear-lanceolate leaves. The inflorescence of P. bamendae is usually longer than leaves and that of P. lindblomii much shorter, up to 3 cm long. Flowers of both species are more or less pubescent with prominent mentum; however, they are easily distinguishable by the lip structure. The labellum of Polystachya bamendae is as wide as short, the lip lateral lobes are prominent up to 2.5 mm long and 2 mm wide, the middle lobe is transversely widely oblong, 4-lobed, emarginate with a prominent apex on the back side and the callus is fleshy bilobed ridge along the midnerve in the middle of the lip. On the contrary the lip of Polystachya lindblomii is twice longer than wide, the lateral lobes are small up to 0.6 mm long and 1 mm wide, the middle lobe is broadly ovate to orbicular and the lip callus is a transverse tridentate fleshy ridge.
The change of frameshift and an occurrence of the stop codon as the consequence are caused by a presence of indel 19 bp within the sequence of matK gene of the species of Superpositae (excluding Polystachya lindblomii) and in the newly described P. bamendae as well. This character is a molecular synapomorphy and along with morphological characters they prove a common origin of the representatives of Superpositae. Russell et al. (2010b) suggest that the matK gene of the mentioned taxa is a pseudogene. However, considering its function in post-transcriptional modification process, matK is essential for proper organism working. Barthet and Hilu (2007) basing on an analysis of matK cDNA translation did not find the stop codons in Spathoglottis plicata Blume (Orchidaceae), which were noted during the DNA analysis of this gene (Freudenstein et al. 2004). An analogical situation may concern the supposed pseudogene in Superpositae. Two alternative hypotheses supporting functionality of the matK gene in the mentioned species may be presented: (1) RNA editing mechanisms can improve the reading frame in species with frameshift indels and premature stop codons; (2) downstream frameshift by one base pair eliminate all stop codons in matK gene in all mentioned species. In this case ATA codon (for isoleucine) could be initial one. ATA and ATT (both codons for isoleucine) were reported as start codon for Thalassiosira pseudonana Cleve and Phaeodactylum tricornutum Bohlin (Chromalveolata), respectively (Imanian et al. 2010).
The question is whether the taxa possessing the duplication are the monophyletic group? The problem is interesting, because Polystachya lindblomii, apart from all other species of the section Superpositae, does not possess the duplication and possessing the indel seems to be a new character. If we had more samples of different representatives of Polystachya londblomii, would the indel appear? We had an access to a relatively small amount of samples and therefore we are not able to unequivocally state it is a constant character, it may be a result of variability within a population or a molecular synapomorphy as well. A hypothesis is the duplication of this fragment took place in the ancestor of all species possessing today the duplication, what makes the group monophyletic under condition that Polystachya londblomii is no longer classified within the group. What is surprising, the new species is the most closely related to P. lindblomii. The analysis of morphological characters of P. lindblomii and other species of the section does not support the hypothesis. To solve the problem and determine the status of indel (synapomophy or not), an access to more samples and results of population studies are needed.
The subsectional division proposed by Mytnik-Ejsmont (2011) does not correlate with the results of the present molecular analyses. Only the nominal subsection is monophyletic, whereas representatives of the rest two subsections (Polystachya spatella and P. kermesina) form strongly supported monophyletic clade. The newly described Polystachya bamendae forms a weakly supporting clade with P. lindblomii (subsection Spatellae), however, in the analysis of the ITS marker only. The plastid marker does not support such relation between the two species. However, it is noteworthy that only a few species were analyzed, less than 50 percent of the representatives of the subsections Spatellae and Superpositae. Therefore, basing on such weakly representing test, there is too early in our opinion to verify the subsectional division proposed by Mytnik-Ejsmont (2011). Undoubtedly, we may state that Polystachya lindblomii is an isolated species within the section.
As most species of the section Superpositae present restricted distribution, they should be subjected to careful conservation action. These species are very fragile in their ecosystems and can easily be driven to extinction (Munishi et al. 2004). At this time we do not have enough information to discuss the conservation status of Polystachya bamendae due to the paucity of the population. It is known so far from a single locality in Cameroon and a field survey needs to be carried out to ascertain the distribution range of the species and propose the IUCN category.
References
Barthet M, Hilu KW (2007) Expression of matK: functional and evolutionary implications. Am J Bot 94(8):1402–1412
BirdLife International (2010) www.birdlife.org. Accessed 25 December 2012
Conservation International (2005) Biodiversity hotspots. www.conservation.org. Accessed 25 December 2012
Cribb PJ (1979) New or little known orchids from East Africa. Kew Bull 34:321–340
Cribb PJ (1984) Orchidaceae 2. In: Polhill RM (ed) Flora of tropical East Africa. Balkema, Rotterdam, pp 237–412
Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB and matK DNA sequences. Am J Bot 89:132–144
Douzery EJP, Pridgeon AM, Kores P, Linder HP, Kurzweil H, Chase MW (1999) Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. Am J Bot 86:887–899
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimal change for a specific tree topology. Syst Zool 20:406–416
Freudenstein J, van den Berg VC, Goldman DH, Kores PJ, Molvray M, Chase MW (2004) An expanded plastid DNA phylogenetic analysis of Orchidaceae and analysis of jackknife clade support strategy. Am J Bot 91:149–157
Galtier N, Gouy M, Gautier C (1996) SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548
Imanian B, Pombert J-K, Keeling PJ (2010) The complete plastid genomes of the two ‘Dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLOS One 5(5):e10711
Kraenzlin F (1926) Monographie der Gattung Polystachya Hook. Repert Spec Nov Regni Veg Beih 39:1–136
Lovett JC (1993) Eastern Arc moist forest flora. In: Lovett JC, Wasser SK (eds) Biogeography and ecology of the rain forests of Eastern Africa. Cambridge University Press, Cambridge, pp 33–57
Molvray M, Kores PJ, Chase MW (2000) Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA (eds) Monocots: Systematics and Evolution. Victoria, CSIRO Publishing, Australia, pp 441–448
Munishi PKT, Shear TH, Wentworth T, Temu RPC, Maliondo SM (2004) Sparse distribution pattern of some plant species in two afromontane rain forests of the Eastern Arc Mountains in Tanzania. Tanzania J For Nat Conserv 75:74–90
Mytnik-Ejsmont J (2011) A monograph of the subtribe Polystachyinae Schltr. (Orchidaceae). Gdańsk, Fundacja Rozwoju Uniwersytetu Gdańskiego
Russell A, Samuel R, Rupp B, Barfuss MHJ, Safran M, Besendorfer V, Chase MW (2010a) Phylogenetics and cytology of a pantropical orchid genus Polystachya (Polystachyinae, Vandeae, Orchidaceae): evidence from plastid DNA sequence data. Taxon 59:389–404
Russell A, Samuel R, Klejna V, Barfuss MHJ, Rupp B, Chase MW (2010b) Reticulate evolution in diploid and tetraploid species of Polystachya (Orchidaceae) as shown by plastid DNA sequences and low-copy nuclear genes. Ann Bot 106:37–56
Swofford DL (2000) PAUP* version 4.0b10. Phylogenetic analysis using parsimony (*and other methods), v. 4.0 beta 10. Sunderland, Sinauer
Thiers B (2008, continuously updated) Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/ih/. Accessed 25 December 2012
WWF (2012) WWF: Terrestrial Ecoregions. www.worldwildlife.org. Accessed 25 December 2012
Acknowledgments
We wish to thank the curators and staff of the cited herbaria for their kind hospitality and assistance during our visits (B, BM, BR, BRLU, C, HBG, K, L, MA, MO, P, W and WAG). We are very grateful to prof. Tomasz Osiejuk and his research team (Adam Mickiewicz University, Poland) for their invaluable help during our field work in Cameroon. We would also like to express our gratitude to Anton Russell for an access to his DNA sequences and stimulating discussions. We are also obliged to an anonymous reviewer for his comments and improving the manuscript. This study was supported by the grants from the Ministry of Science and Higher Education (N N303 343735 and 8124/B/PO1/2011/40). This article was also prepared thanks to the grants from European Commission’s Research Infrastructure Action via the SYNTHESYS Project at the Real Jardin Botanico (CSIC; ESTAF 4129), Natural History Museum of Vienna (AT-TAF 1690, AT-TAF 3903), National Botanic Garden of Belgium (BE-TAF 750), National Herbarium of The Netherlands (NL-TAF 2834, NL-TAF 2265), Natural History Museum of Denmark (DK-TAF 3231, DK-TAF 4040), and Botanic Garden and Botanic Museum in Berlin-Dahlem (DE-TAF 3906).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Mytnik-Ejsmont, J., Szlachetko, D.L., Baranow, P. et al. A phylogenetic and morphological study of Polystachya sect. Superpositae (Orchidaceae) with description of a new species from Cameroon. Plant Syst Evol 300, 19–28 (2014). https://doi.org/10.1007/s00606-013-0856-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0856-0