Abstract
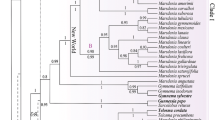
The genus Panus and many of its species have a wide geographic distribution, and in-depth up-to-date taxonomic review is needed that includes critical review of type materials within a phylogenetic frame. In order to recover the phylogenetic relationships within Panus species and their morphological boundaries and to critically analyze the diversity recorded for Brazil, we carried out fieldwork in poorly explored areas in the country and morphological and literature revisions of fungarium specimens, including several type materials. We present a comprehensive phylogeny of Panus and discuss several taxonomic and nomenclatural implications in order to achieve stability for species of the genus. Four new species are proposed, P. capelariae, P. pachysporus, P. speciosus, and P. stiptonotatus. Panus campinensis and P. thailandicus (an endophytic species) are proposed as new combinations in the genus, based on a morphological revision and phylogenetic evidence of their types, respectively. Additionally, Endopandanicola is synonymized within Panus, and P. parvus is synonymized within P. strigellus. The occurrence of P. conchatus, P. convivalis, P. fulvus, P. similis, and P. tephroleucus in Brazil is rejected due to morphological and phylogenetic evidences. For P. conchatus and P. similis, we present bases for the recognition of its sensu stricto status. We also discuss nomenclatural issues surrounding the Lentinus velutinus complex that include the basionym elucidation, its sensu stricto delimitation, and an epitypification based on a new sequenced specimen from the type locality. Our comprehensive assessment of Panus in Brazil has led to the confirmation of ten species supported by morphological and/or molecular data, which are critically discussed, and an identification key is presented.













Similar content being viewed by others
Availability of data and material
All data used in this study are either directly cited (nomenclature) or are available through the cited references (underlying phylogeny and sequence data) or through the cited repositories (MycoBank registration numbers, molecular data at Dataverse).
References
Batista AC, Falcão RGS, Peres GEP, Moura NR (1966) Fungi Paraenses (Revisão da Coleção de Paul C. Hennings, do Museu Paraense Emílio Goeldi). Inst Micol 506:10–290
Berkeley MA (1843) Notices of fungi in the herbarium of the British Museum. Ann Mag Nat Hist 10:369–385
Berkeley MJ (1854) Decades of fungi. Decades XLI- XLIII. Indian fungi. Hooker’s J Bot Kew Gard Miscellany 6:129–143
Berkeley MJ, Broome CE (1873) Enumeration of the fungi of Ceylon. Part II., containing the remainder of the Hymenomycetes, with the remaining established tribes of fungi. J Linn Soc, Bot 14(73):29–140. https://doi.org/10.1111/j.1095-8339.1873.tb00301.x
Berkeley MJ, Curtis MA (1869) Fungi Cubenses (Hymenomycetes). J Linn Soc Bot 10:280–392
Bulliard P (1792) Herbier de la France; ou, Collection complette des plantes indigenes de ce royaume; avec leurs propriétés, et leurs usages en medecine, vol 12. Bulliard, Didot, Debure, and Belin, Paris
Cavalcante FSA, Campos MCC, de Lima JPS (2021) New occurrences of macrofungi (Basidiomycota) in southern Amazonas, Brazil. Ci e Nat 43:e46. https://doi.org/10.5902/2179460X44026
Corner EJH (1981) The agaric genera Lentinus, Panus and Pleurotus with particular reference to Malaysian species. Beih Nova Hedw 69:1–169
Coutinho AXP (1925) Florae mycologicae Insulae St. Thomae. Anais do Instituto Superior de Agronomia da Universidade Técnica de Lisboa, vol 2. Instituto Superior de Agronomia, Lisboa, pp 1–29
Dayarathne MC, Boonmee S, Braun U, Crous PW, Daranagama DA, Dissanayake AJ, Ekanayaka H, Jayawardena R, Jones EB, Maharachchikumbura SS, Perera RH (2016) Taxonomic utility of old names in current fungal classification and nomenclature: Conflicts, confusion & clarifications. Mycosphere 7:1622–1648. https://doi.org/10.5943/mycosphere/7/11/2
Douanla-Meli C, Langer E (2010) Reassessment of phylogenetic species relationship of some lentinoid fungi with velutinate basidiomes based on partial 28S ribosomal RNA gene sequencing. Sydowia 62(1):23–35
Drechsler-Santos ER, Wartchow F, Coimbra VRM, Gibertoni TB, Cavalcanti MAQ (2012) Studies on lentinoid fungi (Lentinus and Panus) from the semi-arid region of Brazil. J Torrey Bot 139:437–446
Ediriweera SS, Nanayakkara CM, Weerasena OV, Karunarathna SC, Wijesundera RL, Piyatissa MA (2021) Morphology and phylogeny reveal nine new records of polypores from dry zone of Sri Lanka. Chiang Mai J Sci 48(3):893–908
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Fries EM (1821) Systema mycologicum, sistens fungorum ordines, genera et species, vol 1. Sumptibus Ernesti Mauritii, Gryphiswaldia, Gryphiswaldiae, p 520
Fries EM (1825) Systema orbis vegetabilis. Primas lineas novae constructionis periclitatur Elias Fries. Pars I. Plantae homonemeae. Typographia Academica, Lund, pp 369
Fries EM (1830) Eclogae fungorum, praecipue ex herbarus germanorum de scriptorum. Linnaea 5:497–553
Fries E (1838) Epicrisis systematis mycologici seu synopsis Hymenomycetum. Typographia Academica, Uppsala. https://doi.org/10.1080/00222934009512452
Galvão VIP, Koroiva R, Wartchow F (2023) A new species of Panus (Panaceae, Polyporales) from Paraíba, Brazil. Phytotaxa 514(3):17. https://doi.org/10.11646/phytotaxa.619.2.5
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Góes-Neto A, Loguercio-Leite C, Guerrero RT (2005) DNA extraction from frozen field-collected and dehydrated herbarium fungal basidiomata: performance of SDS and CTAB-based methods. Biotemas 18(2):19–32
Grand EA (2004) Systematics and species concepts in the genera Lentinus Fr. and Panus Fr., with emphasis on the Lentinus tigrinus, L. crinitus and Panus lecomtei complexes. Thesis, University of Tennessee
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. https://doi.org/10.1093/sysbio/syq010
Hennings P (1897) Fungi camerunenses II. Botanische Jahrbücher für Systematik Pflanzengeschichte und Pflanzengeographie 23:537-558
Hibbett DS, Vilgalys R (1993) Phylogenetic relationships of Lentinus (Basidiomycotina) inferred from molecular and morphological characters. Syst Bot 18:409–433. https://doi.org/10.2307/2419417
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42(2):182–192. https://doi.org/10.2307/2992540
Huelsenbeck JP, Rannala B (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol 53(6):904–913. https://doi.org/10.1080/10635150490522629
Justo A, Miettinen O, Floudas D, Ortiz-Santana B, Sjökvist E, Lindner D, Nakasone K, Niemelä T, Larsson KH, Ryvarden L, Hibbett DS (2017) A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol 121(9):798–824. https://doi.org/10.1016/j.funbio.2017.05.010
Kalchbrenner C (1881) Fungi Macowaniani. Grevillea 9(52):131–137
Katoh S (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. https://doi.org/10.1093/bioinformatics%2Fbts199
Kumar TA, Manimohan P (2005) A new species of Lentinus from India. Mycotaxon 92:119–123
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol and Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Küppers H (1994) Atlas de los colores. Naturart, Barcelona
Lanfear R, Calcott B, Ho SY, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol and Evol 29(6):1695–1701. https://doi.org/10.1093/molbev/mss020
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol and Evol 34(3):772–773. https://doi.org/10.1093/molbev/msw260
Largent DL, Johnson D, Watling R (1977) How to identify mushrooms to genus III: microscopic features. Mad River Press, Eureka, California, p 148
Léveillé JH (1844) Champignons exotiques. Ann Nat Sci, Bot Ser 3(2):167–221
Liu S, Shen LL, Wang Y, Xu TM, Gates G, Cui BK (2021) Species diversity and molecular phylogeny of Cyanosporus (Polyporales, Basidiomycota). Front Microbiol 12:631166. https://doi.org/10.3389/fmicb.2021.631166
Maia LC et al (2015) Diversity of Brazilian Fungi. Rodriguésia 66(4):1033–1045. https://doi.org/10.1590/2175-7860201566407
Meijer AAR (2006) Preliminary list of the macromycetes from the Brazilian State of Paraná. Bol Mus Bot Mun 68:1–59
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). IEEE Press, New Orleans, Louisiana. https://doi.org/10.1109/GCE.2010.5676129
Motato-Vásquez V, Gugliotta AM, Rajchenberg M, Catania M, Urcelay C, Robledo G (2020) New insights on Bjerkandera (Phanerochaetaceae, Polyporales) in the Neotropics with description of Bjerkandera albocinerea based on morphological and molecular evidence. Plant Ecol Evol 153(2):229–245. https://doi.org/10.5091/plecevo.2020.1667
Murrill WA (1915) (Agaricales) Polyporaceae. North Amer Fl 9:201–296
Oliveira-Filho AT (2015) Um sistema de classificação fisionômico-ecológico da vegetação neotropical: segunda aproximação. In: Eisenlohr PV, Felfili JM, de Melo MMRF, de Andrade LA, Meira-Neto JAA (eds) Fitossociologia no Brasil: métodos e estudos de casos, v2. UFV, Viçosa, pp 385−411
Olou BA, Krah FS, Piepenbring M, Yorou NS, Langer E (2020) Diversity of Trametes (Polyporales, Basidiomycota) in tropical Benin and description of new species Trametes parvispora. MycoKeys 65:25. https://doi.org/10.3897/mycokeys.65.47574
Overholts LO (1930) Eu-Basidiomycetes. In: Chardon CE, BA Toro. Mycological explorations of Colombia. J Agric Univ P R 14:195−353. https://doi.org/10.46429/jaupr.v14i4.14223
Palacio M, Robledo GL, Reck MA, Grassi E, Góes-Neto A, Drechsler-Santos ER (2017) Decrypting the Polyporus dictyopus complex: recovery of Atroporus Ryvarden and segregation of Neodictyopus gen. nov. (Polyporales, Basidiomycota). PLoS ONE 12(10):0186183. https://doi.org/10.1371/journal.pone.0186183
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A (2009) How many bootstrap replicates are necessary?. In: Batzoglou, S. (eds) Research in Computational Molecular Biology. RECOMB 2009. Lecture Notes in Computer Science(LNBI), vol 5541. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-02008-7_13
Pegler DN (1971) Lentinus Fr. and related genera from Congo-Kinshasa (Fungi). Bull Jard Bot Natl Belg 41:273–281
Pegler DN (1972) Lentineae (Polyporaceae), Schizophyllaceae et especes lentinoides et pleurotoides des Tricholomataceae. Fl Illust Champ Afr Centre Fasc 1:1–26
Pegler DN (1983) The genus Lentinus: a world monograph. Kew Bull Addit Ser 10:1–281
Pegler DN (1997) The Agarics of São Paulo, Brazil: an account of the agaricoid fungi (Holobasidiomycetes) of São Paulo State, Brazil. Royal Botanic Gardens, UK
Peintner U, Kuhnert-Finkernagel R, Wille V, Biasioli F, Shiryaev A, Perini C (2019) How to resolve cryptic species of polypores: an example in Fomes. IMA Fungus 10:1–21. https://doi.org/10.1186/s43008-019-0016-4
Psurtseva NV, Zmitrovich IV, Seelan JS, Bulakh EM, Hughes KW, Petersen RH (2021) New data on morphology, physiology, and geographical distribution of Lignomyces vetlinianus, its identity with Lentinus pilososquamulosus, and sufficient phylogenetic distance from Le. martianoffianus. Mycol Progress 20:809–821. https://doi.org/10.1007/s11557-021-01701-z
Putzke J (1994) Lista dos fungos Agaricales (Hymenomycetes, Basidiomycotina) referidos para o Brasil. Cad Pesq Univ Fed Santa Cruz Do Sul, Ser Bot 6:3–186
Putzke J, Putzke MTL (2002) Os reinos dos fungos, vol 2. Santa Cruz do Sul, Edunisc, p 212
Raithelhuber J (1974) Hongos argentinos I. Compañía Impresora Argentina, Buenos Aires
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67(5):901–4
Rick J (1907) Fungi austro-americani Fasc. V U VI in: Ann Mycol 5:28–31
Rick J (1930) Contributio ad Monographiam Polyporacearum et Agaricacearum Brasiliensium IV. Brotéria, Sér Bot 24:27–118
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. https://doi.org/10.1093/sysbio/sys029
Saccardo PA (1887) Sylloge Fungorum Omnium Hucusque Cognitorum, vol. V. R. Friedländer & Sohn, Berlin. https://doi.org/10.5962/bhl.title.5371
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sanuma OI, Tokimoto K, Sanuma C, Autuori J, Sanuma LR, Martins MS, Junior NM, Ishikawa NK, Apiamö RM (2016) Sanöma samakönö sama tökö nii pewö oa wi ĩ tökö waheta: Ana amopö= Enciclopédia dos Alimentos Yanomami (Sanöma): Cogumelos. Hutukara Associação Yanomami e Instituto Socioambiental, São Paulo, pp 108
Seynes J de (1897) Recherches pour servir à l’Histoire Naturelle et à la Flore des Champignons du Congo Français, vol 1. Masson & Cie, Paris
Soltis PS, Soltis DE (2003) Applying the bootstrap in phylogeny reconstruction. Statist Sci 18(2):256–267. https://doi.org/10.1214/ss/1063994980
Sousa-Guimarães DK, Alves-Silva G, Camacho O, Menolli Jr N, Góes-Neto A, Souza JF, Robledo RL, Neves MA, Drechsler-Santos ER (2022) Data for: Studies on Panus (Panaceae, Polyporales): morphology and phylogeny assist new species descriptions. Harvard Dataverse, V2. 10.7910/DVN/DZVFKL
Spegazzini C (1889) Fungi Puiggariani: Pugillus I. Bol Acad Nac Ci Cordoba 11:1–381
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1312. https://doi.org/10.1093/bioinformatics/btu033
Stevenson G (1964) The Agaricales of New Zealand V. Kew Bull 19(1):1–59
Teixeira AR (1946) Himenomicetos brasileiros III. Bragantia 6:165–188
Teixeira AR (1995) Método para estudo das hifas do basidiocarpo de fungos poliporáceos. Manual nº 6, Instituto de Botánica, São Paulo, pp 22
Thiers B [continuously updated] Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. Home page at: http://sweetgum.nybg.org/ih/
Tibpromma S et al (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261. https://doi.org/10.1007/s13225-017-0378-0
Tibpromma S, Hyde KD, Bhat JD, Mortimer PE, Xu J, Promputtha I, Doilom M, Yang JB, Tang AMC, Karunarathna SC (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33:25–67. https://doi.org/10.3897/mycokeys.33.23670
Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Kusber WH, Li DZ, Marhold K, May TW, McNeill J, Monro AM, Prado J, Price MJ, Smith GF (2018) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Koeltz Botanical Books, Glashütten. https://doi.org/10.12705/Code.2018
Vargas-Isla R, Capelari M, Meloni N, Nagasawa E, Tokimoto K, Ishikawa NK (2015) Relationship between Panus lecomtei and P. strigellus inferred from their morphological, molecular and biological characteristics. Mycoscience 56(6):561–571. https://doi.org/10.1016/j.myc.2015.05.004
Vellinga EC, Noordeloos ME (2001) Glossary. In: Noordeloos ME, Kuyper ThW, Vellinga EC (eds) Flora agaricina neerlandica, vol 5. CRC Press, Boca Raton, pp 6–11
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Vinjusha N, Kumar TA (2022) Validation of Panus bambusinus and P. roseus (Panaceae, Polyporales). Phytotaxa 533(4):235–236. https://doi.org/10.11646/phytotaxa.533.4.7
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, New York, pp 315–322
Zmitrovich IV, Kovalenko AE (2016) Lentinoid and polyporoid fungi, two generic conglomerates containing important medicinal mushrooms in molecular perspective. Int J Med Mushrooms 18:23–38. https://doi.org/10.1615/intjmedmushrooms.v18.i1.40
Zmitrovich IV, Malysheva VF (2013) Towards a phylogeny of Trametes alliance (Basidiomycota, Polyporales). Mikol Fitopatol 47(6):358–380
Acknowledgements
We are grateful to all the protected areas and their directors for permission to sample collections; the curators of mentioned fungaria for the loan of specimens (FLOR, IAC, SP, URM); the fungaria curatorship that readily sent photos and support us with type specimen data (BAFC, ISC, LISU, LPS, K, TENN, UPS); the Laboratório Multiusuário de Estudos em Biologia (LAMEB/UFSC) for providing infrastructure to carry out the molecular studies; J. Prado, C. Bicudo, and F. Wartchow for nomenclatural discussion on Panus neostrigosus versus Panus lecomtei; Dra. Fernanda Karstedt for pictures of Panus ciliatus; Dra. Viviane de Oliveira Garcia and colleagues from MICOLAB-UFSC for specimen collections; Dr. Diogo Henrique Costa-Rezende and Kelmer Cunha for assistance in analyses and discussion of species and pictures, respectively; and Dr. Jaya Seelan Sathiya Seelan (Univ. of Malaysia Sabah) for generating ITS and nrLSU sequences from some Brazilian specimens loaned from the herbarium SP. This research is part of the MIND.Funga research group: https://mindfunga.ufsc.br/.
Funding
The authors thank Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES) for providing master and PhD scholarships to DKSG and FB; the PPGFAP/UFSC; the FONCYT (PICT 0830 to GR) and Fundación FungiCosmos for partial financing of the research; the Society of Systematic Biologists for the Mini-ARTS award to FB, allowing type revisions at LPS. NMJr. thanks the “Fundação de Amparo à Pesquisa do Estado de São Paulo” (FAPESP) (grant #18/15677–0). AGN, ERDS, NMJr., and GAS are supported by CNPq (Grant Nos. 308880/2022–6, 310150/2022–1, 314236/2021–0, and 153025/2022–0, respectively).
Author information
Authors and Affiliations
Contributions
Conceptualization: Denyse K. Sousa-Guimarães, Elisandro R. Drechsler-Santos; methodology: Denyse K. Sousa-Guimarães, Felipe Bittencourt; formal analysis and investigation: Denyse K. Sousa-Guimarães, Olga Camacho, Genivaldo Alves-Silva, Elisandro R. Drechsler-Santos; writing—original draft preparation: Denyse K. Sousa-Guimarães, Genivaldo Alves-Silva, Elisandro R. Drechsler-Santos; writing—review and editing: Felipe Bittencourt, Gerardo L. Robledo, Nelson Menolli Jr, Aristóteles Góes-Neto; funding acquisition: Elisandro R. Drechsler-Santos, Aristóteles Góes-Neto; resources: Elisandro R. Drechsler-Santos, Aristóteles Góes-Neto; supervision: Genivaldo Alves-Silva, Nelson Menolli Jr, Gerardo L. Robledo; Elisandro R. Drechsler-Santos.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Zhu-Liang Yang
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sousa-Guimarães, D.K., Alves-Silva, G., Bittencourt, F. et al. A comprehensive phylogeny of Panus (Panaceae, Polyporales) and revisited Brazilian diversity. Mycol Progress 23, 19 (2024). https://doi.org/10.1007/s11557-024-01955-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-024-01955-3