Abstract
We investigated genetic variation of 273 individuals from 25 populations of the monotypic species Hagenia abyssinica (Rosaceae) from the highlands of Ethiopia at three chloroplast microsatellite loci. The objectives were to infer the factors that shaped the genetic structure and to reconstruct the recolonization history of the species. Six haplotypes that were phylogenetically grouped into two lineages were identified. Homology of the three loci to the respective regions of the chloroplast genome was confirmed by sequencing. The chloroplast haplotypes found in Hagenia showed a clear pattern of congruence between their geographical distribution and genealogical relationships. A very low haplotype diversity within populations (h S = 0.079, v S = 0.058) and a very high population differentiation (G ST = 0.899, N ST = 0.926) was observed, reflecting very low mixing between recolonizing lineages. Restricted gene flow through seeds, rare long-distance dispersal, contiguous range expansion and mutation shaped the genetic structure of Hagenia. Fossil pollen records suggested that the trend of postglacial recolonization of Hagenia was first in the south and latter went to the north in Ethiopia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic variation is structured not only by the contemporary forces of genetic exchange but also by historical patterns of relationship (Schaal et al. 1998). Phylogeographic analyses can provide insights into the historical processes responsible for restricted distributions of populations (Cruzan and Templeton 2000). Phylogeography characterizes population subdivision by recognizing geographical patterns of genealogical structure across the range of a species (Avise 2004), synthesizing the influence of both history and current genetic exchange (Schaal et al. 1998). Cladistic gene genealogies can form the basis of historical approaches to the study of intraspecific processes (Schaal et al. 1998; Templeton et al. 1987; Templeton 2004). Rare long-distance gene flow events potentially have great evolutionary significance (Le Corre et al. 1997; Schaal et al. 1998; Cain et al. 2000) and great biological relevance by shaping genetic variation (Ouborg et al. 1999). Rare dispersal events produce fragmented advancing fronts establishing new populations as a result of dispersal from pioneer populations, as well as from populations that are part of the continuous distribution (Cruzan and Templeton 2000). Cain and co-workers (2000) argued that rare events can control the rate of population spread and that only dispersal via seed directly affects colonization of new populations. For plant populations that have passed through recent episodes of range expansion, long-distance dispersal events are probably the most important factors of spatial genetic structuring at maternally inherited genes at small or medium geographic scales (Le Corre et al. 1997). In the simulation-based study on colonization dynamics of maternally inherited loci in oak, Le Corre and co-workers (1997) demonstrated that stratified dispersal was far more rapid than pure diffusion, even if long-distance dispersals were very rare events. They also argued that long-distance dispersal events influenced the genetic differentiation of populations, leaving a genetic signature that is likely to persist for long periods. This paper demonstrates the significance of such rare events, among others, in shaping the genetic structure of the monotypic species Hagenia abyssinica.
Chloroplast DNA has been widely used in the investigations of genetic structure (e.g. Meister et al. 2005; Parducci et al. 2001), phylogeography (e.g. Butaud et al. 2005; Meister et al. 2005; King and Ferris 1998; Rendell and Ennos 2002) and colonization history (e.g. Cavers et al. 2003; Petit et al. 2002, 2003; Heuertz et al. 2004) of tree species. We employed chloroplast microsatellite markers (Weising and Gardner 1999) to investigate genetic structure, phylogeography and recolonization routes of H. abyssinica within its natural range in Ethiopia. The mode of inheritance of the chloroplast genome of H. abyssinica has not been determined, but it is most likely maternally inherited as in the majority of angiosperms (Harris and Ingram 1991; Birky 1995; Finkeldey and Hattemer 2007). We also examined some fossil pollen records in order to examine the recolonization history of Hagenia after late Pleistocene ice ages in Africa.
Hagenia is a wind-pollinated and wind-dispersed broad-leaved dioecious tree species that belongs to a monotypic genus in the Rosaceae family (Hedberg 1989; Negash 1995). It is confined to Africa and its ecological range stretches from Ethiopia in the North to Zimbabwe in the South (Hedberg 1989; http://www.worldagroforestry.org). Fossil pollen records indicated that Hagenia recolonized Ethiopia from the south during the late Pleistocene and became abundant in the southern regions of Ethiopia about 2,500 years Before Present (BP) (Hamilton 1982; Umer 1992; Umer et al. 1996; Umer and Bonnefille 1998; Umer et al. 2007). At present, the extant Hagenia populations throughout the country are situated at higher altitudes, often in wetter depressions.
The tree has remarkably diversified economic and ecological values (Azene et al. 1993; Berhanu et al. 1999; Dawit and Ahadu 1993; Jansen 1981; Hedberg 1989). Hagenia has been logged heavily and selectively and it is one of the endangered tree species in Ethiopia (Negash 1995).
Genetic inventories of H. abyssinica are rare and restricted only to some parts of the species’ distribution range. Kumilign (2005) and Feyissa et al. (2007) investigated the genetic diversity of a few populations of H. abyssinica using anonymous RAPD and ISSR markers, respectively. The present investigation using cpDNA is the first of its kind and covered the whole range of the species in Ethiopia. We predicted that (1) since the seeds of H. abyssinica are dispersed by wind, there is low differentiation among populations but high variation within populations, (2) populations show geographic structuring primarily induced by mutation and isolation by distance, (3) based on the existing fossil pollen records, Hagenia recolonized Ethiopia after the Last Glacial Maximum (LGM) more particularly becoming abundant during the late Holocene in contrast to the southern regions. For the latter the recolonization was much earlier.
Two main questions are addressed: (1) which factors shaped the maternally inherited genetic variation of Hagenia in Ethiopia? (2) Is there a congruence between molecular data and palynological evidences to infer the relationships among genealogical lineages and migration routes of the species?
Materials and methods
Sampling and DNA extraction
Twenty-two natural and three planted populations were sampled from all regions where Hagenia is known to grow in Ethiopia. These populations represent most of the extant distribution of the species in the country. The distribution of the populations stretches from 05°51′N (Hagere-Mariam) in the south to 13°11′N (Debark-Mariam) in the north, and from 35°42′E (Wonbera) in the west to 40°14′E (Dindin) in the east (Table 1; Fig. 3). The distances between populations range from 21 to 806 km. The populations are located within an altitudinal range of 2,200 m a.s.l. at Bonga to 3,200 m a.s.l. at Wofwasha, and temperatures range from an absolute minimum of −1°C at Dinsho to a maximum of 33.5°C at Kosso Ber populations. The nearest meteorological stations are situated at lower altitudes than Hagenia populations in most of the cases, and therefore, higher rainfall and lower temperatures are expected than those shown in Table 1.
Young leaves were collected and partially desiccated in paper bags before drying with silica gel and stored at room temperature before DNA extraction. Genomic DNA was isolated from leaves following the DNeasy 96 kit protocol of Qiagen® (Hilden, Germany). In an initial test, DNA was isolated from dried leaves of different sizes. A size of 1 cm2 (about 20 mg) gave the best results with regard to DNA quantity and quality and was used for all samples.
PCR amplification and genotyping
The 10 pairs of consensus chloroplast microsatellite primers (CCMPs) (Weising and Gardner 1999) were tested on three samples from three geographically separated populations that are far from each other. Seven of them gave amplification products, three of which (CCMP2, CCMP6 and CCMP10) were found to be polymorphic, and were used to screen 273 samples (9–12 individuals from each population). Additional 144 samples were analyzed to study spatial genetic structure in four polymorphic natural populations. DNA was diluted (1:10) prior to PCR amplification. PCR reactions were performed in a Peltier Thermal Cycler PTC-200 (MJ Research®), with a volume of 16 μl reaction mixture containing 2 μl HPLC H2O, 8 μl hot star master mix (containing 10 mM Tris–HCL (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM each of dNTPs, 0.8U Taq DNA polymerase) (Qiagen®, Hilden, Germany), 2 μl of each forward and reverse primer (5 pmol/μl) and 2 μl DNA (about 10 ng). The forward primer was labeled with the fluorescent dyes 6-FAM or HEX. The PCR profile for CCMP2 and CCMP10 was 15 min initial denaturation at 95°C, followed by 35 cycles of 1 min denaturation at 94°C, 1 min annealing at 50°C and 1 min extension at 72°C, with a final extension of 10 min at 72°C. The PCR profile for CCMP6 differed in the annealing temperature (52.5°C). Aliquots of the amplification products were diluted prior to electrophoretic separation on the ABI 3100 Genetic Analyser (Applied Biosystems®) depending on the intensity of the bands observed after agarose gel electrophoresis. Two microliters diluted (multiplexed in most cases) PCR product were denaturated for 2 min at 90°C with 12 μl HiDi formamide (Applied Biosystems®) containing ~0.02 μl internal size standard (GS ROX 500, Applied Biosystems®) before loading on the ABI Genetic Analyser 3100 (Applied Biosystems®) for separation.
Sequencing
Comparative sequencing of 18 fragments from the three chloroplast loci was performed to confirm the amplified regions and to determine the molecular basis for size variation. The amplification products were purified using the QIAquick Gel Extraction kit (Qiagen®, Germany) following the manufacturer’s specifications. We employed direct sequencing for a locus having relatively larger fragment sizes, CCMP2 (224–235 bp). Cloning was performed for the two loci with smaller fragment sizes, CCMP6 (140–142 bp) and CCMP10 (96–97 bp), using the pBSKS vector and X BlueI competitive cells with the TA cloning method (Invitrogen®). Sequencing followed the dideoxy-chain termination method (Sanger et al. 1977). Sequencing reaction of 10 μl total volume containing 1 μl Big Dye (BD vers. 3.1), 1.5 μl sequencing buffer (SB 3.1), 4.8 μl HPLC H2O, 0.7 μl forward or reverse primer (5 pmol/μl), 2 μl purified DNA (about 10 ng) was used. Since no sequences of CCMP2 from other species of the family Rosaceae were available in existing databases, three out-group species from the Rosaceae family that were available in the Botanic Garden of the Georg-August-University Göttingen, Germany, were also sequenced for comparison. The sequence data have been stored in the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) with the accession numbers FM174367-75 and FM 174387 for locus CCMP2 (10 sequences), FM174376-80 for locus CCMP6 (5 sequences), FM174381-83 for locus CCMP10 (3 sequences), and FM1743784-86 for the locus CCMP2 of the out-group species (3 sequences).
Data analysis
Amplification products were aligned with the internal size standard using GENESCAN 3.7, and fragments were scored with GENOTYPER 3.7 (Applied Biosystems®). Polymorphisms in fragment size were identified as different length variants that were combined to define haplotypes. Genetic diversity (h s, h T, v S, v T) and differentiation among populations (G ST, N ST) was computed by PermutcpSSR (available at http://www.pierroton.inra.fr/genetics/labo/Software/PermutCpSSR/index.html) as described by Pons and Petit (1995, 1996). Distribution of genetic diversity within and among populations was estimated by an analysis of molecular variance (AMOVA) using ARLEQUIN Version 3.0 (Excoffier et al. 2005; available at http://cmpg.unibe.ch/software/arlequin3).
Sequences were analyzed with the sequence analyzing software 3.7 (Applied Biosystems®), edited by the program BIOEDIT (Hall 1999) and aligned with Clustal W application (Thompson et al. 1994; available at http://www.ebi.ac.uk/clustalw/).
A statistical parsimony network of haplotypes was constructed with the help of a program TCS Version 1.21 (Clement et al. 2000) from DNA sequence data. Large gaps in a sequence due to an indel (insertion/deletion) are coded as a single mutation to avoid theoretical intermediate haplotypes that are created by the program, which interprets each gap as independent mutation event. The sequence data also confirmed that the larger indel was the result of a single mutation event. The TCS program was also used to compute the out-group weights of haplotypes. A nested clade phylogeographic analysis (NCPA) of the spatial distribution of the genetic variation was performed by the program GEODIS (Posada et al. 2000). Nested clades were plotted manually on the haplotype network based on the algorithms defined by Templeton et al. (1987). The interpretation of statistically significant patterns of distribution was made following the inference key described in Templeton (2004).
Results
Genetic diversity and differentiation
We found three alleles in locus CCMP2, three alleles in locus CCMP6 and two alleles in locus CCMP10 (Tables 2 and 3). The analyses of within population diversity (h S), total diversity (h T) and differentiation (G ST) yielded 0.079, 0.787 and 0.899, respectively, under the assumption of unordered haplotypes (Pons and Petit 1996). The corresponding values for within population diversity (v S), total diversity (v T) and differentiation (N ST) with ordered haplotypes (Pons and Petit 1996), taking genetic distances among haplotypes into account, were 0.058, 0.787 and 0.926, respectively. An AMOVA showed that 92.3% of the total genetic diversity is represented among populations. A test of spatial genetic structure in the four polymorphic natural populations did not show any family or spatial genetic structure, indicating effective seed dispersal by wind at the local level. This is not unexpected for species with very light wind-dispersed seeds. The additional 144 individuals analyzed for spatial genetic structure showed similar haplotype composition and frequencies, as the sample that was analyzed to study genetic diversity (Table 4). No additional haplotypes were found due to increased sample size.
Results of sequencing
Multiple sequence alignments of loci CCMP2, CCMP6 and CCMP10 are shown in Fig. 1. Sequencing confirmed homology of the three loci to the respective regions of the chloroplast genome. The observed variations were due to variable numbers of poly (A) or poly (T) repeats and a large indel of 10 bp at position 100 bp in the flanking region of the locus CCMP2. In total, there were four variable sites (3 short indels in the microsatellites and a large indel in the flanking region). A 10-bp segment is preceded by an identical sequence in the flanking region of the four genotypes in locus CCMP2 (underlined in Fig. 1). The duplication can be explained by a strand slippage during cpDNA replication in a single mutational event (e.g. Wolfson et al. 1991). The sequences of the three out-group species (Rubus fruticosus, R. idaeus and Rosa canina) also showed duplication events of different segments in similar region (Fig. 1).
Sections of sequences of three Hagenia chloroplast microsatellite loci (a CCMP2 aligned with Rubus fruticosus, Rubus idaeus and Rosa canina, b CCMP6, c CCMP10). Duplications in locus CCMP2 are underlined. Microsatellite repeats are shown in bold. Gaps indicate deletions of nucleotides. i individual; f fragment size. Identical sequences from different individuals are not shown
Phylogeography
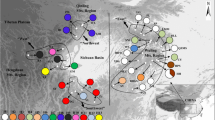
Six haplotypes (H1–H6) were identified from the combination of the three loci as detected by fragment analysis (Table 3). A fully resolved statistical parsimony network of the chloroplast haplotypes of Hagenia, which is reconstructed from DNA sequences (Fig. 2), demonstrates the relationship among the different haplotypes and the minimum number of evolutionary events separating them. The third frequent haplotype, H4 (represented in 21% of the individuals), has a larger out-group weight (0.35) than the most frequent haplotype H1 (represented in 34% of the individuals) with an out-group weight of 0.26. Out-group weight is a relative weight of haplotypes based on mutational steps and is strongly correlated with actual age (of a haplotype) and thus is a much better indicator of haplotype age than is the haplotype frequency (Castelloe and Templeton 1994). All of the haplotypes are separated by a single mutation step. H1 is separated from H4 by a deletion of 10 nucleotides in locus CCMP2.
Statistical parsimony network showing nested clades and relatedness among haplotypes of Hagenia abyssinica at three chloroplast loci. Size of circles are proportional to their respective out-group weights. Thick bar indicates indel of 10 bp in a single mutation event; thin bar indicates indel of 1 bp
The observed N ST value is significantly higher than the G ST value at p<0.01 (none of the permutated G ST values was higher than the observed N ST value), indicating geographical clustering of related haplotypes. Three nested clades were evident from the haplotype network (Fig. 2) and the χ 2 statistic revealed a significant association (p<0.0001) between genealogical and geographic distributions in all of the clades (Table 5). Restricted gene flow was inferred for the haplotypes nested in clade 1-1 (but with some long-distance dispersal events over intermediate areas) and in clade 1-2 (with isolation by distance), while contiguous range expansion was deduced from the total cladogram (Table 5).
Haplotype H1 is widely distributed in central Ethiopia, while H2 is common in southern regions (Fig. 3). Haplotype H4 has the longest geographic distribution in south-north direction stretching from the southwest to the northern regions. The northern population DK was established most likely by single long-distance dispersal event. A recent mutation at locus CCMP10 resulted in the rarest haplotype (H5) that is restricted to only one population (Wonbera) in the west (fixed on one type), distinguishing it from other populations. Haplotype H6 is restricted to the central-northern region, while H3 has only a rare occurrence in the southern region, and is always in association with H2 and/or H1. The domination of the population Uraga (UR) by haplotype H3 as contrasting to the neighboring populations (H2) in the south was caused by a single mutational event in locus CCMP6.
Geographic distribution of the chloroplast haplotypes of Hagenia abyssinica in Ethiopia. Dotted enclosure shows lineage I; dashed enclosure shows lineage II. Grey dashed lines indicate approximate position of the Great Rift Valley. The inset pie chart shows the relative frequency distribution of the haplotypes. Two letters designate populations as in Table 1; H1–H6 indicate haplotypes as in Table 3. The arrows indicate the putative recolonization route of the species. Brighter shaded areas show extant closed forests; darker shaded areas show lakes. Source map: Assefa (unpublished)
The three loci exhibited different impacts on haplotypic variation in the different regions of the country. Locus CCMP6 accounted for the variation in central and southern Ethiopia, while CCMP2 was responsible for the variation in southwestern and northern regions. Locus CCMP10 caused the variation in West Ethiopia due to the prevalence of a private allele. Eighty percent of the populations are fixed on one haplotype, while the remaining populations share two to three haplotypes. Two populations (KL and BR) contained three similar haplotypes at different frequencies while three populations (UR, CM and KDP) possessed two different haplotypes each. Two of the planted populations included in this study (DKP and SMP) showed haplotypes that were identical to those of their respective parent populations (DK and SM) based on the record obtained from the District Office of Agriculture (unpublished data). The source population of a third plantation (KDP) was not confirmed, but it exhibited a combination of haplotypes from two neighboring populations (H1 and H6), suggesting that seeds were obtained from the adjacent populations or alternatively, they were procured from the national seed center.
Discussion
Genetic diversity and differentiation
A very low genetic and haplotype diversity within populations (h S = 0.079, v S = 0.058, respectively) and a very high population differentiation (G ST = 0.899, N ST = 0.926) proved a marked genetic separation of the populations. This result rejects our first prediction that there is low differentiation among populations, but high variation within populations. The population differentiation is much higher in the chloroplast genome than in the nuclear genome of H. abyssinica, as revealed by ISSR (G ST = 0.25; Feyissa et al. 2007) and AFLP markers (G ST = 0.15, Ayele et al. submitted).
Likewise, Rendell and Ennos (2002) found a population differentiation that was 10-fold higher in the chloroplast genome of Calluna vulgaris (L.) Hull (Ericaceae), than in the nuclear genome. In general, maternally inherited genomes experienced considerably more subdivision (mean G ST value of ~0.64) than biparentally inherited genomes (mean G ST value of ~0.18) of angiosperm species (reviewed by Petit et al. 2005). The coefficient of population differentiation (G ST) in Hagenia is higher than or comparable to G ST values recorded for other species with heterogeneous mode of seed dispersal, including wind dispersal, investigated by chloroplast markers (Newton et al. 1999; Petit et al. 2003).
The high level of genetic variation among populations of Hagenia suggested a restricted migration of seeds among regions, which is also reflected in the observed geographic structuring of haplotypes. The demographic history of the species and/or existence of natural barriers (mountains, valleys and long distances) to seed dispersal might account for the strong phylogeographic pattern. Young and Boyle (2000) reported that for wind-pollinated and dispersed species, the pattern of gene flow and genetic structure is a function of interfragment distance. Though Hagenia is a montane species, its migration is not necessarily along the mountains as evidenced by haplotype H1 that is distributed at both sides of the Great Rift Valley (Fig. 3), most likely due to long-distance seed dispersal as intermediate populations are missing.
Phylogeographical and palynological interpretation
The geographic distribution of haplotypes (Fig. 3) and their genealogical relationships (Fig. 2) observed in Hagenia demonstrated a marked phylogeographical structure as a result of highly restricted gene flow via seeds. Such patterns arise when scattering is reduced because the novel mutations remain localized within the geographical context of their origins (e.g. Butaud et al. 2005). Both the G ST-N ST test and the NCPA detected a very strong association between genealogical and geographic distributions. The NCPA inferred that restricted gene flow associated with contiguous range expansion shaped the genetic structure of Hagenia. This result allows us to accept the second prediction that populations show geographic structuring primarily induced by isolation by distance, coupled with local mutation events. Two distinct lineages that were separated by an indel of 10 nucleotides in locus CCMP2 are evident from the cladogram (Fig. 2). The first lineage constitutes haplotype H4 and its derived haplotypes H5 and H6 that are distributed in the southwestern and northern regions (referred hereafter as lineage I), while the second lineage embodies H1 and its derived haplotypes H2 and H3 in central and southern regions (lineage II). Such a non-random distribution of haplotypes asserts our prediction on the existence of phylogeographic pattern in Hagenia.
In light of the palynological records discussed at the end of this section, there are two possible scenarios for the immigration of Hagenia into Ethiopia. The first scenario suggests that the lineage I of Hagenia recolonized Ethiopia first through the southwestern mountains (population Bonga, BG) that is situated to the west of the Great Rift Valley whereas the second scenario suggests that lineage II of Hagenia recolonized Ethiopia first through the southern mountains (population Hagere-Mariam, HM) situated east of the Great Rift Valley. However, our data supports the first scenario. The cladogram demonstrated that haplotype H4 is the most probable ancient haplotype that served as a root for the rest of the haplotypes because of its higher out-group weight. Castelloe and Templeton (1994) argued that the most ancient haplotype should be located at the center of the gene tree and be geographically widespread, whereas the most recent haplotypes should be at the tips of the gene tree and be localized geographically. This ascertains the postulation that haplotype H4 is the most ancient haplotype (followed by H1), whereas H2, H3, H5 and H6 are located at the tips of the gene tree and are highly localized geographically. This observation suggests that Hagenia recolonized Ethiopia most likely through the southwestern mountains of the country (population Bonga, BG) from southern African regions and expanded and diversified in the central, southern and northern regions of the country. Our results demonstrated that lineage I most likely gave rise to lineage II due to a deletion of 10 nucleotides in a single mutational event. This mutational event was quite recent assuming the recent recolonization of Ethiopia by Hagenia. All the southern haplotypes were most likely derived from a single seed parent with haplotype H4 and expanded to the central regions and diversified into the southern regions. In general, our results confirm that recolonization took place only from the south. The inferred recolonization routes and putative long-distance dispersal event are shown in Fig. 3.
The long gap observed between the northern and southern populations with the haplotype H4 led to three postulations: (1) the populations that are situated between these two regions diversified to other haplotypes due to mutation (e.g. population WB diversified to H5 and population DR diversified to H6, both based on a single mutation event), (2) some populations containing the same haplotype might be lost due to anthropogenic activities, (3) natural or human mediated colonization events in terms of long-distance seed dispersal or purposeful seed transfer account for this disjunct haplotype distribution. The postulations, however, are not exclusive. Haplotype H1 is also widely distributed, and such a widespread distribution of individual haplotypes indicates rapid range expansion (Schaal et al. 1998) and the significant role of rare events, particularly long-distance seed dispersal and mutation, in shaping the genetic diversity in Hagenia. Furthermore, the patchy structure of the haplotypes of Hagenia, in general, is a result of rare long-distance dispersal of seeds during colonization, each patch resulting from a founding event beyond the colonizing front.
Climate conditions over the LGM show dry conditions over East Africa, when lake levels were lower than today or even dried out (Lamb et al. 2007; Umer et al. 2004). The LGM was dry and cold on the mountains (Umer et al. 2004). For example, Lake Tana, source of the Blue Nile, in northern Ethiopia was desiccated between 16,700 and 15,100 cal BP (Lamb et al. 2007). These areas today receive mean annual rainfall of nearly 1,400 mm/year. In the south western part of the country mean annual rainfall today reaches 2,000 mm. Therefore, it can be supposed that one area for the LGM refugium could likely be located in the south western part of the country. This implies that the recent recolonization of Hagenia in the northern uplands is the result of climatic amelioration after the LGM. Moreover, fossil pollen data shows that signals of postglacial Hagenia expansion started earlier in the southern regions (Burundi, Uganda, Kenya) (Hamilton 1982; Bonnefille et al. 1995; Jolly et al. 1997) when compared with the Ethiopian mountains where it was much later (Hamilton 1982; Umer and Bonnefille 1998; Umer et al. 2007). While the refugium for upland tree species including Hagenia is not known so far, they might have recolonized the northern highlands out of either regions mentioned above. Though complete coverage is unavailable from Africa, the palynological data obtained from fossil pollen stratigraphy of some sites (Table 6) suggested that the postglacial recolonization of Hagenia followed a northward route with the oldest available record from Burundi (ca. 34,000 14C yrs BP). Major expansion of Hagenia took place around 11,500 14C yrs BP in Burundi (Bonnefille et al. 1995) and Uganda, such as in the area of Ahakagyezi swamp (Jolly et al. 1997). Its major expansion on the Bale Mountains (southern Ethiopia) was after 2,500 cal yrs BP (Umer et al. 2004; Umer and Bonnefille 1998). Bonnefille and Umer (1994) also reported that Hagenia expanded after 590 yrs BP in Arsi Mountains (central Ethiopia). In general, the signal of Hagenia in the pollen records in Ethiopia was quite high in the late Holocene epoch and the palynological evidences in general suggested a possible northward migration route also within Ethiopia. The fossil pollen records support our third prediction that Hagenia recolonized Ethiopia after the LGM more particularly becoming abundant during the late Holocene in contrast to the southern regions. The examination of the same pollen diagrams that are described above also indicated a northward recolonization of some other tree species such as Podocarpus falcatus, Juniperus procera and Olea species in Africa. The palynological data also showed that the fossil pollen accumulation of H. abyssinica has been alarmingly declining through time in the African countries other than Ethiopia, suggesting a sequential reduction in the size of the populations.
It remains uncertain from the pollen data whether there was a forest refugium during the glacial/late glacial period at lower altitudes than 2,000–2,500 m in southern Ethiopia. However, Holocene pollen diagrams from such areas as the Rift Valley do not suggest that the refugium was there (Lezine and Bonnefille 1982; Lamb et al. 2000). The reason could be that LGM climate was too dry to support forest refugia. Southwestern Ethiopia could be a possible refugium as supported by the molecular data and current climatic conditions.
Conclusions
The joint interpretation of genealogical relationships among the chloroplast haplotypes and the fossil pollen evidences allowed us to accept the hypothesis predicting recolonization of Ethiopia by Hagenia after the LGM from the south of Ethiopia and possibly south western Ethiopia. The first is supported by palynological information whereas the second is by molecular and current climate information. There was no indication of past fragmentation of Hagenia populations from our results, pointing to the effect of random long-distance seed dispersal. It is most likely that populations were established from few parent seed trees. Given the mountainous topography of the country that is intermittently dissected by wide valleys, Hagenia did not have a continuous distribution. The cpDNA assay detected sufficient variation for a phylogeographic study of H. abyssinica in Ethiopia. A remarkable subdivision of cpDNA diversity in the species was found, as indicated by a high level of genetic differentiation. The chloroplast haplotypes of H. abyssinica demonstrated a pattern of isolation-by-distance. Unlike most of the wind-dispersed tree species, the chloroplast haplotypes found in Hagenia showed a clear pattern of congruence between their geographical distribution and genealogical relationships, allowing us to accept the prediction on geographic structuring. Analysis of cpDNA types and palynological inventories, including all countries where the species is known to grow, would fully resolve the genealogical relationships and help to identify the glacial refugia of Hagenia in Africa. The analysis of pollen records from different sites and altitudes in Ethiopia where Hagenia is growing would help to fully understand the recolonization route of the species within the country.
References
Abegaz BM, Ngadjui BT, Bezabih M, Mdee LK (1999) Novel natural products from marketed plants of eastern and southern Africa. Pure Appl Chem 71:919–926
Avise JC (2004) Molecular markers, natural history and evolution, 2nd edn. Sinauer Associates Inc, Sunderland
Azene BT, Birnie A, Tegnäs B (1993) Useful trees and shrubs for Ethiopia: identification, propagation and management for agricultural and pastoral communities. Technical Handbook No. 5, Regional Conservation Unit, Swedish International Development Authority (SIDA)
Beuning KRM, Talbot MR, Kelts K (1997) A revised 30,000-year paleoclimatic and paleohydrologic history of Lake Albert, East Africa. Palaeogeogr Palaeocl 136:259–279
Birky CW Jr (1995) Uniparental inheritance of mitochondria and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA 92:11331–11338
Bonnefille R, Umer MM (1994) Pollen-inferred climatic fluctuations in Ethiopia during the last 3000 years. Palaeogeogr Palaeocl 109:331–343
Bonnefille R, Riollet G, Buchet G, Icole M, Lafont R, Arnold M, Jolly D (1995) Glacial/interglacial record from intertropical Africa, high resolution pollen and carbon data at Rusaka, Burundi. Quaternary Sci Rev 14:917–936
Butaud J-F, Rives F, Verhaegen D, Bouver J-M (2005) Phylogeography of Eastern Polynesian sandalwood (Santalum insulare), an endangered tree species from the pacific: a study based on chloroplast microsatellites. J Biogeogr 32:1763–1774
Cain ML, Milligan BG, Strand AE (2000) Long-distance seed dispersal in plant populations. Am J Bot 87:1217–1227
Castelloe J, Templeton AR (1994) Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol 3:102–113
Cavers S, Navarro C, Lowe AJ (2003) Chloroplast DNA phylogeography reveals colonization history of a Neotropical tree, Cedrela odorata L., in Mesoamerica. Mol Ecol 12:1451–1460
Clement M, Posada D, Grandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cruzan MB, Templeton AR (2000) Paleoecology and coalescence: phylogeographic analysis of hypotheses from the fossil record. Trends Ecol Evol 15:491–496
Darbyshire I, Lamb H, Umer MM (2003) Forest clearance and regrowth in northern Ethiopia during the last 3000 years. Holocene 13:537–546
Dawit A, Ahadu A (1993) Medicinal plants and enigmatic health practices of Northern Ethiopia. Berhanena Selam Printing Press, Addis Ababa
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Feyissa F, Nybom H, Bartish IV, Welander M (2007) Analysis of genetic diversity in the endangered tropical tree species Hagenia abyssinica using ISSR markers. Genet Resour Crop Evol 54:947–958
Finkeldey R, Hattemer HH (2007) Tropical forest genetics. Springer, Berlin
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98
Hamilton AC (1982) Environmental history of East Africa. A study of the quaternary. Academic Press, London
Harris SA, Ingram R (1991) Chloroplast DNA and biosystematics: the effects of intraspecific diversity and plastid transmission. Taxon 40:393–412
Hedberg O (1989) Rosaceae. In: Hedberg I, Edwards S (eds) Flora of Ethiopia, Vol. 3, Pittosporaceae to Arralaceae. The National Herbarium, Addis Ababa
Heuertz M, Fineschi S, Anzidei MA et al (2004) Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol Ecol 13:3437–3452
Jansen PCM (1981) Spices, condiments and medicinal plants in Ethiopia, their taxonomy and agricultural significance. Agricultural Research Reports 906. Center for Agricultural Publishing and Documentation, Wageningen
Jolly D, Taylor D, Marchant R, Hamilton A, Bonnefille R, Buchet G, Riollet G (1997) Vegetation dynamics in central Africa since 18,000 yr BP: pollen records from the interlacustrine highlands of Burundi, Rwanda and western Uganda. J Biogeogr 24:495–512
King RA, Ferris C (1998) Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Mol Ecol 7:1151–1163
Kumilign A (2005) Estimation of sex-related genetic diversity of Hagenia abyssinica (Bruce) J.F. Gmel. M.Sc. thesis, Addis Ababa University
Lamb HF (2001) Multi-proxy records of Holocene climate and vegetation change from Ethiopian Crater lakes. Proy Irish Acad B 101:35–46
Lamb AL, Leng MJ, Lamb HF, Umer MM (2000) A 10,000-year oxygen and carbon isotope record of hydrological change in a small Ethiopian crater lake. The Holocene 10:167–177
Lamb AL, Leng MJ, Umer MM, Lamb HF (2004) Holocene climate and vegetation change in the main Ethiopian Rift Valley, inferred from the composition (C/N and δ13C) of lacustrine organic matter. Quaternary Sci Rev 23:881–891
Lamb HF, Bates CR, Coombes PV, Marshall MH, Umer MM, Davies SJ, Dejen E (2007) Late Pleistocene desiccation of Lake Tana, source of the Blue Nile. Quaternary Sci Rev 26:287–299
Le Corre V, Machon N, Petit RJ, Kremer A (1997) Colonization with long-distance seed dispersal and genetic structure of maternally inherited genes in forest trees: a simulation study. Genet Res 69:117–125
Lezine AM, Bonnefille R (1982) Diagramme Pollinique Holocene d’un sondage du lac Abiyata (Ethiopie, 7°42′ Nord). Pollen et Spore 3–4:463–480
Meister J, Hubaishan MA, Killian N, Oberprieler C (2005) Chloroplast DNA variation in the shrub Justicia areysiana (Acanthaceae) endemic to the monsoon affected coastal mountains of the southern Arabian Peninsula. Bot J Linn Soc 148:437–444
Negash L (1995) Indigenous trees of Ethiopia: biology, uses and propagation techniques. SLU, Reprocentralen, Umeå
Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Olago DO, Street-Perrott FA, Perrott RA, Ivanovich M, Harkness DD (1999) Late quaternary glacial-interglacial cycle of climatic and environmental change on Mount Kenya. J Afr Earth Sci 29:593–618
Ouborg NJ, Piquot Y, Van Groenendael JM (1999) Population genetics, molecular markers and the study of dispersal in plants. J Ecol 87:551–568
Parducci L, Szmidt AE, Madaghiele A, Anzidei M, Vendramin GG (2001) Genetic variation at chloroplast microsatellites (cpSSRs) in Abies nebrodensis (Lojac.) Mattei and three neighboring Abies species. Theor Appl Genet 102:733–740
Petit RJ, Brewer S, Bordács S et al (2002) Identification of refugia and post-glacial colonization routes of European white oaks based on chloroplast DNA and fossil pollen evidence. Forest Ecol Manag 156:49–74
Petit RJ, Aguinagalde I, de Beaulieu J-L et al (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Pons O, Petit RJ (1995) Estimation, variance and optimal sampling of gene diversity. 1. Haploid locus. Theor Appl Genet 90:462–470
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall KA, Templeton AR (2000) GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol Ecol 9:487–488
Rendell S, Ennos RA (2002) Chloroplast DNA diversity in Calluna vulgaris (heather) populations in Europe. Mol Ecol 11:69–78
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Templeton AR (2004) Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol Ecol 13:789–809
Templeton AR, Boerwinkle E, Sing CF (1987) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117:343–351
Thompson JD, Higgins DG, Gibson TG (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Umer MM (1992) Paléoenvironment et paléoclimatologie des derniers millenaires en Ethiopie, contribution palynologique. PhD Thesis, Univ. ďAix Marseille 3
Umer MM, Bonnefille R (1991) The recent history of vegetation and climate around lake Langano (Ethiopia). Paleoecol Afr 22:275–286
Umer MM, Bonnefille R (1998) A Late Glacial to Late Holocene pollen record from a high land peat at Tamasaa, Bale mountains, South Ethiopia. Global Planet Change 16–17:121–129
Umer MM, Bonnefille R, Johnson TC (1996) Pollen and isotopic records of late Holocene sediments from Lake Turkana, N. Kenya. Palaeogeogr Palaeocl 119:371–383
Umer MM, Dagnachew N, Gasse F, Bonnefille R, Lamb HF, Leng MJ (2004) Late quaternary climate changes in the Horn of Africa. In: Battarbee RW, Gasse F, Stickly CE (eds) Past climate variability through Europe and Africa. Kluwer, Dordrecht, pp 159–177
Umer MM, Lamb HF, Bonnefille R, Lézine A-M, Tiercelin J-J, Gibert E, Gazet J-P, Watrin J (2007) Late Pleistocene and Holocene vegetation history of the Bale Mountains, Ethiopia. Quaternary Sci Rev 26:2229–2246
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Wolfson R, Higgins KG, Sears BB (1991) Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol Biol Evol 8:709–721
Young AG, Boyle TJ (2000) Forest fragmentation. In: Young A, Boshier D, Boyle T (eds) Forest conservation genetics: principles and practice. CABI Publishing, UK
Acknowledgments
This work is supported by the German Federal Ministry of Economic Cooperation and Development (BMZ) through the German Technical Cooperation (gtz) as a component of a project “Support to the Forest Genetic Resources Conservation Project” of the Ethiopian Institute of Biodiversity Conservation (IBC). The German Academic Exchange Service (DAAD) executed the grant as a PhD project of the first author. The National Meteorological Service Agency of Ethiopia provided climatic data free of charge. We are indebted to Oleksandra Dolynska and Thomas Seliger for technical assistance in the laboratory.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ayele, T.B., Gailing, O., Umer, M. et al. Chloroplast DNA haplotype diversity and postglacial recolonization of Hagenia abyssinica (Bruce) J.F. Gmel. in Ethiopia. Plant Syst Evol 280, 175–185 (2009). https://doi.org/10.1007/s00606-009-0177-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-009-0177-5