Abstract
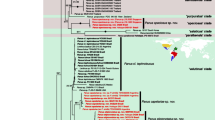
All known Italian Gagea species (23) representing seven out of 14 sections were included in this study. Morphological, molecular and combined data support the basal position of G. trinervia (Anthericoides). According to the molecular data, the sections Anthericoides, Lloydia, Gagea, Minimae and Spathaceae are very well supported; representatives of the sections Didymobulbos and Fistulosae clustered together in the same clade. In this clade, several near-related species groups (series) were recognised. According to 35 morpho-anatomical, ontogenetic and karyological data the section Gagea forms a well-defined monophyletic clade. Within this clade G. tisoniana, G. pusilla and G. cfr. paczoskii are near related. The position of the critical putative hybrid taxa G. luberonensis and G. polidorii is discussed. New series Solenarium (Dulac) Peruzzi and Tison, Occidentales (A. Terracc.) Peruzzi and Tison and Saxatiles (A. Terracc.) Peruzzi and Tison are proposed.





Similar content being viewed by others
References
Ali SI (2006) Two new species of Gagea Salisb. (Liliaceae) from Pakistan. Pak J Bot 38(1):43–46
Allen GA, Soltis DE, Soltis PA (2003) Phylogeny and biogeography of Erythronium (Liliaceae) inferred from chloroplast matK and nuclear rDNA ITS sequences. Syst Bot 28:512–523
Bartolucci F, Peruzzi L (2008) Distribuzione del genere Gagea Salisb. (Liliaceae) nell’Appennino centro-settentrionale. Biogeographia (in press)
Bayer E, López González G (1989) Nomenclatural notes on some names in Gagea Salisb. (Liliaceae). Taxon 38(4):643–645
Bohdanowicz J, Lewandowska B (1999) Participation of endoplasmic reticulum in the unequal distribution of plastids during generative cell formation in Gagea lutea (L.) Ker-Gawl. (Liliaceae). Acta Biol Crac Ser Bot 41:177–183
Caparelli KF, Peruzzi L, Cesca G (2006) A comparative analysis of embryo-sac development in three closely-related Gagea Salisb. species (Liliaceae), with some consideration on their reproductive strategies. Pl Biosyst 140(2):115–122
Cuccuini P, Luccioli E (1995) Tipificazione di Ornithogalum spathaceum Hayne (Liliaceae) e presenza di Gagea spathacea (Hayne) Salisb. nella flora italiana. Webbia 49(2):253–264
Davlianidze MT (1972) Generis Gagea Salisb. conspectus specierum Caucasicarum I. Notulae Systematicae ac Geographicae Instituti Botanici Thbilissensis 29:69–75
Eickbush TH, Eickbush DG (2007) Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics 175:477–485
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ferrer Gallego PP, Laguna Lumbreras E, Alba Villegas S, Tison JM (2007) Sobre la presencia de Gagea lacaitae A. Terracc. (Liliaceae) en la flora valenciana. Acta Bot Malacitana 32:1–12
Gargano D, Peruzzi L, Caparelli KF, Cesca G (2007) Preliminary observations on the reproductive strategies in five early-flowering species of Gagea Salisb. (Liliaceae). Bocconea 21:349–358
Greilhuber J, Ebert I, Lorenz A, Vyskot B (2000) Origin of facultative heterochromatin in the endosperm of Gagea lutea (Liliaceae). Protoplasma 212:217–226
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Henker H (2005) Die Goldsterne von Mecklenburg-Vorpommern unter besonderer Berücksichtigung kritischer und neuer Sippen. Botanischer Rundbrief für Mecklenburg-Vorpommern 39:5–89
Huelsenbeck JP, Ronquist F (2001) Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol 51:673–688
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Molec Evol 16:111–120
Ikinci N, Oberprieler C, Güner A (2006) On the origin of European lilies: phylogenetic analysis of Lilium section Liriotypus (Liliaceae) using sequences of the nuclear ribosomal transcribed spacers. Willdenowia 36:647–656
John H, Peterson A, Peterson J (2004) Zum taxonomischen Rang zweier kritischer Sippen der Gattung Gagea in Mitteleuropa. Mitt. florist. Kart. Sachsen-Anhalt (Halle) 9:15–26
Levichev IG (1990) The synopsis of the genus Gagea (Liliaceae) from the western Tien-Shan. Bot Zhurn 75(2):225–234
Levichev IG (1999a) Phytogeographical analysis of the genus Gagea Salisb. (Liliaceae). Komarovia 1:47–59
Levichev IG (1999b) The morphology of Gagea Salisb. (Liliaceae) I. Subterranean organs. Flora 194:379–392
Levichev IG (2001) New species of the genus Gagea Salisb. (Liliaceae) from western regions of Asia. Turczaninowia 4(1–2):5–35
Levichev IG (2006a) Four new species of the genus Gagea Salisb. (Liliaceae) from Western Himalayas and the adjoining regions. Pak J Bot 38(1):47–54
Levichev IG (2006b) A review of the Gagea (Liliaceae) species in the flora of Caucasus. Bot Zhurn 91(6):917–951
Levichev IG, Ali SI (2006) Seven new species of the genus Gagea Salisb. (Liliaceae) from Western Himalayas and adjoining regions. Pak J Bot 38(1):55–62
Levichev IG, Tison J-M (2004) Typification of the Caucasian Gagea (Liliaceae) taxa described by Karl Koch. Candollea 59:119–133
Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T (1999) A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J Molec Evol 49:238–249
Patterson TB, Givnish TJ (2002) Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: Insights from rbcL and ndhF sequence data. Evolution 56:233–252
Patterson TB, Givnish TJ (2004) Geographic cohesion, chromosomal evolution, parallel adaptive radiations, and consequent floral adaptations in Calochortus (Calochortaceae): evidence from a cpDNA phylogeny. New Phytol 161:253–264
Peruzzi L (2003) Contribution to the cytotaxonomical knowledge of Gagea Salisb. (Liliaceae) sect. Foliatae A. Terracc. and synthesis of karyological data. Caryologia 56(1):115–128
Peruzzi L (2006) Taxonomic considerations on the nomenclatural types of Gagea rhodiaca A. Terracc. and G. reticulata subsp. africana A. Terracc. (Liliaceae), kept at Pisa (PI). Atti Soc Tosc Sci Nat Mem B 113:69–71
Peruzzi L (2008a) Contribution to the cytotaxonomical knowledge of the genus Gagea Salisb. (Liliaceae). III. New karyological data from the central Mediterranean area. Caryologia 61(1):92–106
Peruzzi L (2008b) Studi biosistematici nel genere Gagea Salisb. (Liliaceae): recenti acquisizioni e problemi ancora aperti. Inform Bot Ital 39(Suppl.1):143–146
Peruzzi L (2008c) Hybridity as a main evolutionary force in the genus Gagea Salisb. (Liliaceae). Pl Biosyst 142(1):179–183
Peruzzi L, Aquaro G (2005) Contribution to the cytotaxonomical knowledge of Gagea Salisb. (Liliaceae) II. Further karyological studies on Italian populations. Candollea 60(1):237–253
Peruzzi L, Bartolucci F (2006) Gagea luberonensis J.-M. Tison (Liliaceae) new for the Italian flora. Webbia 61(1):1–12
Peruzzi L, Bartolucci F, Frignani F, Minutillo F (2007) G. tisoniana, a new species of Gagea Salisb. sect. Gagea (Liliaceae) from c Italy. Bot J Linn Soc 155(3):337–347
Peruzzi L, Caparelli KF (2007) Gagea peduncularis (J. & C. Presl) Pascher (Liliaceae) new for the Italian flora. Webbia 62(2):261–268
Peruzzi L, Gargano D (2005) Distribuzione del genere Gagea Salisb. (Liliaceae) in Calabria. Inform Bot Ital 37(2):1117–1124
Peruzzi L, Tison JM, Peterson A, Peterson J (2008) On the phylogenetic position and taxonomic value of Gagea trinervia (Viv.) Greuter and the whole G. sect. Anthericoides A. Terracc. (Liliaceae). Taxon (in press)
Peruzzi L, Tison J-M (2004) Typification and taxonomic status of eleven taxa of Gagea Salisb. (Liliaceae) described by Achille and Nicola Terracciano and conserved at Napoli (NAP). Candollea 59(2):325–346
Peruzzi L, Tison J-M (2005) Typification and taxonomic status of six taxa of Gagea Salisb. (Liliaceae) described from Sicily and conserved at Palermo (PAL). Candollea 60(2):289–298
Peruzzi L, Tison J-M (2006) Typification of the names and taxonomic status of six taxa of Gagea Salisb. (Liliaceae) conserved at Firenze (FI). Candollea 61(2):293–303
Peruzzi L, Tison J-M (2007) Typification of six critical Mediterranean Gagea Salisb. (Liliaceae) species. Candollea 62(2):173–188
Peruzzi L, Zarrei M (2007) Typification of some critical taxa of Gagea Salisb. (Liliaceae) from W Asia. Candollea 62(2):237–244
Peterson A, John H, Koch E, Peterson J (2004) A molecular phylogeny of the genus Gagea (Liliaceae) in Germany inferred from non-coding chloroplast and nuclear DNA sequences. Pl Syst Evol 245:145–162
Peterson A, Levichev IG, Peterson J (2008) Systematics of Gagea and Lloydia (Liliaceae) and infrageneric classification of Gagea based on molecular and morphological data. Molec Phylogenet Evol 46:446–465
Rešetnik I, Liber Z, Satovic Z, Cigić P, Nikolić T (2007) Molecular phylogeny and systematics of the Lilium carniolicum group (Liliaceae) based on nuclear ITS sequences. Pl Syst Evol 265(1–2):45–58
Rønsted N, Law S, Thornton H, Fay MF, Chase MW (2005) Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium (Liliaceae; Liliales) and the infrageneric classification of Fritillaria. Molec Phylogenet Evol 35:509–527
Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sang T, Crawford DJ, Stuessy TF (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Amer J Bot 84:1120–1136
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0b10. Sinauer Associates, Sunderland
Tan D-Y, Zhang Z, Li X-R, Hong D-Y (2005) Restoration of the genus Amana Honda (Liliaceae) based on a cladistic analysis of morphological characters. Acta Phytotax Sin 43(3):262–270
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tison JM (1998) Note complementaire sur quelques Gagea français. Monde Pl 462:7–8
Tison J-M (2001) Typification de Gagea cossoniana Pascher, de Gagea fragifera (Vill.) Ehr. Bayer et G. Lopez et de Gagea maroccana (A. Terracc.) Sennen et Mauricio. Candollea 56(1):197–202
Tison J-M (2004a) Gagea polidorii J.-M. Tison, espèce méconnue du sud-ouest des Alpes et des Apennins. Acta Bot Gall 151(3):319–326
Tison J-M (2004b) Identité et situation taxonomique de Gagea polymorpha Boissier. Candollea 59(1):109–117
Tison J-M (2004c) Contribution a la connaissance du genre Gagea Salisb. (Liliaceae) en Afrique du Nord. Lagascalia 24:67–87
Tison JM, Perret P (2004) Typification d’Ornithogalum pusillum F. W. Schmidt et relations taxonomiques entre Gagea pusilla (F. W. Schmidt) Sweet, Ornithogalum clusii Tausch et G. clusiana Schult. & Schult. f. Candollea 59(1):103–108
Uphof JCT (1958) A review of the genus Gagea Salisb. Pl Life 14:124–133
Van de Peer Y, De Wachter R (1994) Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Volkov RA, Komarova NY, Hemleben V (2007) Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst Biodivers 5:261–276
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Zander RH (2004) Minimal values for reliability of bootstrap and jackknife proportions, decay index, and Bayesian posterior probability. Phyloinformatics 2:1–13
Zarrei M, Zarre S (2005a) A new species of Gagea (Liliaceae) from Iran. Nord J Bot 23:269–274
Zarrei M, Zarre S (2005b) Pollen morphology of the genus Gagea (Liliaceae) in Iran. Flora 200:96–108
Zarrei M, Zarre S, Wilkin P, Rix M (2007) Systematic revision of the genus Gagea Salisb. (Liliaceae) in Iran. Bot J Linn Soc 154(4):559–588
Zhao LQ, Yang J (2006) Gagea daqingshanensis (Liliaceae), a new species from Inner Mongolia, China. Ann Bot Fenn 43:223–224
Zhao YZ, Zhao LQ (2003) A new species of Gagea (Liliaceae) from Nei Mongol, China. Acta Phytotax Sin 41(4):393–394
Zhao YZ, Zhao LQ (2004) Gagea chinensis (Liliaceae), a new species from Inner Mongolia, China. Ann Bot Fenn 41:297–298
Acknowledgments
We are grateful to Igor Levichev, for providing some plant material for the molecular studies. We would like to thank Doerte Harpke from the Biocentre of the Martin-Luther-University of Halle for her assistance in the Bayesian Analyses and Gianni Bedini from the University of Pisa for useful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peruzzi, L., Peterson, A., Tison, JM. et al. Phylogenetic relationships of Gagea Salisb. (Liliaceae) in Italy, inferred from molecular and morphological data matrices. Plant Syst Evol 276, 219–234 (2008). https://doi.org/10.1007/s00606-008-0081-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-008-0081-4