Abstract
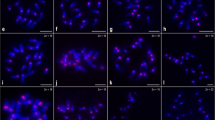
In fifteen geographically isolated populations of five species of Alstroemeria L. (A. aurea, A. hookeri, A. ligtu, A. pelegrina and A. presliana) collected in Chile, karyotypes and variation of RAPD markers were investigated. Tandemly repeated DNA sequences - 5S and 18/25S rDNA genes and the sequence A001-1 (De Jeu et al. 1997) were used to characterize karyotypes by fluorescence in situ hybridization (FISH). Ten somatic metaphases per population were used for measurement of chromosome length. Differences in RAPD marker bands were used for characterization of populations, creating a similarity index. FISH with all three DNA probes shows a high degree of polymorphism between and sometimes also within accessions of A. aurea, A. hookeri and A. ligtu. The number of chromosome pairs showing 5S rDNA signals is more different for the investigated species A. aurea, A. hookeri, A. ligtu, A. pelegrina and A. presliana with 5, 7, 5, 3 and 7, respectively, than the number of 18/25S rDNA signals in this succession with 7, 7, 6, 5 and 7 chromosome pairs, showing a high evolutionary dynamics within the genus. Furthermore, among the four populations of A. hookeri, accession 4181 was different in arm length of chromosome 3. RAPD markers (index of similarity) also showed a greater genetic distance of accession 4181 from the other three accessions of A. hookeri. The possible evolutionary mechanisms providing these polymorphisms were discussed.
Similar content being viewed by others
References
Adachi J, Watanabe K, Fukui K, Ohmido N and Kosuge K (1997). Chromosomal location and reorganization of the 45S and 5S rDNA in the Brachyscome lineariloba complex (Asteraceae). J Pl Res 110: 371–377
Arano H and Saito H (1980). Cytological studies in family Umbelliferae 5. Karyotypes of seven species in subtribe Seselinae. Kromosomo 2: 471–480
Avramova Z (2002). Heterochromatin in animals and plants. Similarities and differences. Pl Physiol 129: 40–49
Bennett S, Leitch I and Bennett M (1995). Chromosome identification and mapping in the grass Zingeria biebersteiniana (2n = 4) using fluorochromes. Chrom Res 3: 101–108
Buitendijk J and Ramanna M (1996). Giemsa C-banded karyotypes of eight species of Alstroemeria L. and some of their hybrids. Ann Bot 78: 449–457
Buitendijk J, Boon E and Ramanna M (1997). Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot 79: 343–353
Buitendijk J, Peters A, Quene R and Ramanna M (1998). Genome size variation and C-band polymorphism in Alstroemeria aurea, A. ligtu and A. magnifica (Alstroemeriaceae). Pl Syst Evol 212: 87–106
Cuadrado A, Ceoloni C and Jouve N (1995). Variation in highly repetitive DNA composition of heterochromatin in rye studied by fluorescence in situ hybridization. Genome 38: 1061–1069
Cuadrado A and Jouve N (1995). Fluorescent in situ hybridization and C-banding analyses of highly repetitive DNA sequences in the heterochromatin of rye (Secale montanum Guss) and wheat incorporating S. montanum chromosome segments. Genome 38: 795–802
De Jeu M, Laschuit J, Chevalier F and Visser R (1995). Hybrid detection in Alstroemeria by use of species-specific repetitive probes. Acta Horti Orn Pl Imp 420: 62–64
De Jeu M, Lasschuit J, Kuipers A, Kamstra S and Visser R (1997). Characterization and localization of repetitive DNA sequences in the ornamental Alstroemeria aurea Graham. Theor Appl Genet 94: 982–990
De Jong J, Fransz P and Zabel P (1999). High resolution FISH in plants – techniques and applications. Trends Pl Sci 4: 258–263
Dellaporta S, Wood J and Hicks J (1983). A plant DNA minipreparation: Version II. Pl Molec Biol Rep 1: 19–21
Gottlob-McHugh S, Levesque M, MacKenzie K, Olson M, Yarosh O and Johnson D (1990). Organization of the 5S rRNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33: 486–494
Grewal S and Moazed D (2003). Heterochromatin and epigenetic control of gene expression. Science 301: 798–802
Gupta P, Sharma S, Kumar S, Balyan H, Beharav A and Nevo E (2004). Adaptive ribosomal DNA polymorphism in wild barley at a mosaic microsite, Nese Yaàr in Israel. Pl Sci 166: 1555–1563
Hall K and Parker J (1995). Stable chromosome fission associated with rDNA mobility. Chrom Res 3: 417–422
Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen W and Maluszynska J (2006). Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97: 205–216
Houben A, Leach C, Verlin D, Rofe R and Timmis J (1997). A repetitive DNA sequence common to the different B chromosomes of the genus Brachycome. Chromosoma 106: 513–519
Ito M, Miyamoto J, Mori Y, Fujimoto S, Uchiumi T, Abe M, Suzuki A, Tabata S and Fukui K (2000). Genome and chromosome dimensions of Lotus japonicus. J Pl Res 113: 435–442
Kakeda K, Fukui K and Yamagata H (1991). Heterochromatic differentiation in barley chromosomes revealed by C- and N-banding techniques. Theor Appl Genet 81: 144–150
Kamstra S, Ramanna M, De Jeu M, Kuipers A and Jacobsen E (1999). Homoeologous chromosome pairing in the distant hybrid Alstroemeria aurea x A. inodora and the genome composition of its backcross derivatives determined by fluorescence in situ hybridization with species-specific probes. Heredity 82: 69–78
Kamstra S, Kuipers A, De Jeu M, Ramanna M and Jacobsen E (1997). Physical localisation of repetitive DNA sequences in Alstroemeria: karyotyping of two species with species-specific and ribosomal DNA. Genome 40: 652–658
Koo D, Choi H, Cho J, Hur Y and Bang J (2005). A high-resolution karyotype of cucumber (Cucumis sativus L. `Winter Long') revealed by C-banding, pachytene analysis, and RAPD-aided fluorescence in situ hybridization. Genome 48: 534–540
Levan A, Fredga K and Sandberg A (1964). Nomenclature for centromeric poisition on chromosomes. Hereditas 52: 201–220
Lim K, Wennekes J, de Jong F, Jacobsen E and van Tuyl J (2001). Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridisation. Genome 44: 911–918
Neves N, Delgado M, Silva M, Caperta A, Morais-Cecilio L and Viegas W (2005). Ribosomal DNA heterochromatin in plants. Cytogenet Genome Res 109: 104–111
Muñoz M, Moreira A (2003) Alstroemerias de Chile. Diversidad, distribución y conservación. Taller La Era, Santiago. 140 p
Panzera F, Giménez M, López J, Giménez G, Cuadrado A, Shaw P, Beven A, Canovas J and DeLaTorre C (1996). Nucleolar organizer expression in Allium cepa L. chromosomes. Chromosoma 105: 12–19
Pedersen C and Linde-Laursen I (1994). Chromosomal locations of four minor rDNA loci and a marker microsatellite sequence in barley. Chrom Res 2: 65–71
Peterka H, Budahn H, Schrader O, Ahne R and Schűlze W (2004). Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor Appl Genet 109: 30–41
Redi C, Garagna S, Zacharias H, Zuccotti M and Capanna E (2001). The other chromatin. Chromosoma 110: 136–147
Reeves A (2001). MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 44: 439–443
Rieseberg L (2001). Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358
Ruas C, Vanzela A, Santos M, Fregonezi J, Ruas P, Matzenbacher N and de Aguiar-Perecin M (2005). Chromosomal organization and phylogenetic relationships in Hypochaeris species (Asteraceae) from Brazil. Genet Molec Biol 28: 129–139
Sanso A (2002). Chromosome studies in Andean taxa of Alstroemeria (Alstroemeriaceae). Bot J Linn Soc 138: 451–459
Sanso A and Hunziker J (1998). Karyological studies in Alstroemeria and Bomarea (Alstroemeriaceae). Hereditas 129: 67–74
Schmidt T and Heslop-Harrison JS (1998). Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Pl Sci 3: 159–199
Schrader O, Budahn H and Ahne R (2000). Detection of 5S and 25S rRNA genes in Sinapis alba, Raphanus sativus and Brassica napus by double fluorescence in situ hybridization. Theor Appl Genet 100: 665–669
Schubert I and Wobus U (1985). In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92: 143–148
Stephens J, Tsuchiya T and Hughes H (1993). Chromosome studies in Alstroemeria pelegrina L. Int J Pl Sci 154: 565–571
Stevenson M, Armstrong S, Jones G and FordLloyd B (1999). Distribution of a 375 bp repeat sequence in Allium (Alliaceae) as revealed by FISH. Pl Syst Evol 217: 31–42
Stupar R, Song J, Tek A, Cheng Z, Dong F and Jiang J (2002). Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics 162: 1435–1444
Vicari M, Artoni R and Bertollo L (2003). Heterochromatin polymorphism associated with 18S rDNA: a differential pathway among Hoplias malabaricus fish populations. Cytogenet Genome Res 101: 24–28
Wang X, Chen P, Liu D, Zhang P, Zhou B, Friebe B and Gill B (2001). Molecular cytogenetic characterization of Roegneria ciliaris chromosome additions in common wheat. Theor Appl Genet 102: 651–657
Weiss H and Maluszynska J (2000). Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana. Hereditas 133: 255–261
Weiss-Schneeweiss H, Stuessy T, Siljak-Yakovlev S, Baeza C and Parker J (2003). Karyotype evolution in South American species of Hypochaeris (Asteraceae, Lactuceae). Pl Syst Evol 241: 171–184
Williams J, Kubelik A, Livak K, Rafalski J and Tingey S (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18: 6531–6535
Yakura K and Tanifuji S (1983). Molecular cloning and restriction analysis of Eco RI-fragments of Vicia faba rDNA. Pl Cell Physiol 24: 1327–1330
Zhang D and Sang T (1998). Chromosomal structure reaarangement of Paeonia brownii and P. californica revealed by fluorescence in situ hybridization. Genome 41: 848–853
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baeza, C., Schrader, O. & Budahn, H. Characterization of geographically isolated accessions in five Alstroemeria L. species (Chile) using FISH of tandemly repeated DNA sequences and RAPD analysis. Plant Syst. Evol. 269, 1–14 (2007). https://doi.org/10.1007/s00606-007-0591-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-007-0591-5