Abstract
A rapid and accurate detection of pathogens is essential for bedside or on-site diagnosis. Filter paper is an ideal diagnostic tool as it requires no equipment, possesses a high surface-area-to-volume ratio and a high capacity of capillary force. The functionalization of the surface of cellulose filter paper was explored by using glutaric anhydride, N-hydroxysuccinimide, and N,N′-dicyclohexylcarbodiimide. The activated surface systems enable aminated DNA to be immobilized on the surface of filter paper. Both synthetic oligonucleotides and bacterial genomic DNA of Staphylococcus aureus, Escherichia coli, and Campylobacter jejuni were detected successfully. The system produces a clear, consistent and highly visible brown signal within 1–5 min. The digital image can also be analyzed quantitatively due to the brown color resulting from the presence of magnetic beads. Bacterial DNA detection was accomplished by using 16S rDNA probe on the activated paper surface for universal bacterial diagnosis. The method is stable and repeatable. It can detect at least 0.5 pmol of a 120-base synthetic oligonucleotide per assay and 5–10 ng of bacterial DNA per assay.

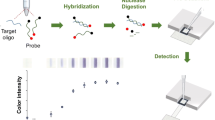
Schematic representation of the method: a. functionalization of cellulose filter paper, b. printing of aminated probes, c. incubation, d. blocking of unreacted functional groups (as dots shown), e. a visual detection of targets, f. quantitative analysis of image.





Similar content being viewed by others
References
Song Y, Gyarmati P (2019) Optimized detection of bacteria in bloodstream infections. PLoS One 14:e0219086. https://doi.org/10.1371/journal.pone.0219086
Ye W, Chen T, Mao Y, Tian F, Sun P, Yang M (2017) The effect of pore size in an ultrasensitive DNA sandwich-hybridization assay for the Escherichia coli O157:H7 gene based on the use of a nanoporous alumina membrane. Microchim Acta 184(12):4435–4844
Chen Y, Wang Z, Liu Y, Wang X, Li Y, Ma P, Gu B, Li H (2018) Recent advances in rapid pathogen detection methods based on biosensors. Eur J Clin Microbial Infect Dis 37:1021–1037. https://doi.org/10.1007/s10096-018-3230-x
Dincer C, Bruch R, Kling A, Dittrich PS, Urban GA (2017) Multiplexed point-of-care testing – xPOCT. Trends Biotechnol 35:728–742. https://doi.org/10.1016/j.tibtech.2017.03.013
Arora P, Sindhu A, Dilbaghi N, Chaudhury A (2011) Biosensors as innovative tools for the detection of food borne pathogens. Biosens Bioelectron 28:1–12. https://doi.org/10.1016/j.bios.2011.06.002
Hol FJ, Dekker C (2014) Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346:1251821. https://doi.org/10.1126/science.1251821
Zhou W, Lie J, Chen Y, Cai Y, Hong Z, Chai Y (2019) Recent advances in the microfluid devices for bacteria and fungus research. TrAC Trends Anal Chem 112:175–195. https://doi.org/10.1016/j.trac.2018.12.024
Chinnadayyala SR, Park J, Le HTN, Santhosh M, Kadam AN, Cho S (2019) Recent advances in microfluidic paper-based electrochemiluminescence analytical devices for point-of-care testing applications. Biosens Bioelectron 126:68–81. https://doi.org/10.1016/j.bios.2018.10.038
Posthuma-Tumpie GA, Korf J, van Amerongen A (2009) Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literaturesurvey. Anal Bioanal Chem 393:569–582. https://doi.org/10.1007/s00216-008-2287-2
Blaskoza M, Koet M, Rauch P, Amerogen AV (2009) Development of a nucleic acid lateral flow immunoassay for simultaneous detection of Listeria spp. and Listeriamonocytogenes in food. Eur Food Res Technol 229:867–874
Carter DJ, Cary RB (2007) Lateral flow microarrays: a novel platform for rapid nucleic acid detection based on miniaturized lateral flow chromatography. Nucleic Acids Res 35:e74. https://doi.org/10.1093/nar/gkm269
Edwards KA, Baeumner EJ (2008) Liposome-enhanced lateral-flow assays for the Sandwich-hybridization detection of RNA. Methods Mol Biol 504:185–215. https://doi.org/10.1007/978-1-60327-569-9_13
Choi JR, Tang R, Wang S, Wan Abas WA, Pingguan-Murphy B, Xu F (2015) Paper-based sample-to-answer molecular diagnostic platform for point-of-care diagnostics. Biosens Bioelectron 74:427–439. https://doi.org/10.1016/j.bios.2015.06.065
Morales-Narvaez E, Naghdi T, Zor E, Merkoci A (2015) Photoluminescent lateral-flow immunoassay revealed by graphene oxide: highly sensitive paper-based pathogen detection. Anal Chem 87:8573–8577. https://doi.org/10.1021/acs.analchem.5b02383
Hu J, Wang L, Li F, Han YL, Lin M, Lu TJ, Xu F (2013) Oligonucleotide-linked gold nanoparticle aggregates for enhanced sensitivity in lateral flow assays. Lab Chip 13:4352–4357. https://doi.org/10.1039/c3lc50672j
Chen Y, Cheng N, Xu Y, Huang K, Luo Y, Xu W (2016) Point-of-care and visual detection of P. aeruginosa and its toxin genes by multiple LAMP and lateral flow nucleic acid biosensor. Biosens Bioelectron 81:317–323. https://doi.org/10.1016/j.bios.2016.03.006
Liu CC, Yeung CY, Chen PH, Yeh MK, Hou SY (2013) Salmonella detection using 16S ribosomal DNA/RNA probe-gold nanoparticles and lateral flow immunoassay. Food Chem 141:2526–2532. https://doi.org/10.1016/j.foodchem.2013.05.089
Song Y (2014) Advances in DNA detection. Dissertation, Royal Institute of Technology (KTH)
Song Y, Gyarmati P, Araujo AC, Lundeberg J, Brumer H 3rd, Stahl PL (2014) Visual detection of DNA on paper chips. Anal Chem 86:1575–1582. https://doi.org/10.1021/ac403196b
Dineva MA, MahiLum-Tapay L, Lee H (2007) Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst 132:1193–1199
Afshari A, Schrenzel J, Jeven M, Harbarth S (2012) Bench-to-bedside review: rapid molecular diagnostics for bloodstream infection – a new frontier? Crit Care 16:222. https://doi.org/10.1186/cc11202
Araújo AC, Song Y, Lundeberg J, Ståhl PL, Brumer H 3rd (2012) Activated paper surfaces for the rapid hybridization of DNA through capillary transport. Anal Chem 84:3311–3317. https://doi.org/10.1021/ac300025v
Benters R, Niemeyer CM, Drutschmann D, Blohm D, Wöhrle D (2002) DNA microarrays with PAMAM dendritic linker systems. Nucleic Acids Res 30:E10
Talbo MJ, White RG (2013) Methanol fixation of plant tissue for scanning Electron microscopy improves preservation of tissue morphology and dimensions. Plant Methods 9:36. https://doi.org/10.1186/1746-4811-9-36
Poulsen L, Soe MJ, Snakeneborg D, Moller LB, Dufva M (2008) Multi-stringency wash of partially hybridized 60-mer probes reveals that the stringency along the probe decreases with distance from the microarray surface. Nucleic Acids Res 36:E132. https://doi.org/10.1093/nar/gkn600
Savage MD (1996) An introduction to avidin-biotin technology and options for biotinylation. In: Meier T, Fahernholz F (eds) A laboratory guide to biotin-labeling in biomolecule analysis. Birkhauser Verlag, Basel, p 11
Ravan H, Kashanian S, Sanadgol N, Badoei-Dalfard A, Karami Z (2014) Strategies for optimizing DNA hybridization on surfaces. Anal Biochem 444:41–46
Xiao W, Huang J (2011) Immobilization of oligonucleotides onto zirconia-modified filter paper and specific molecularrecognition. Langmuir 27:12284–12288. https://doi.org/10.1021/la203150f
Weiss LR, Orzel RA (1967) Some comparative toxicologic and pharmacologic effects of dimethyl sulfide as a pesticide solvent. Toxicol Appl Pharmacol 11:546–557
Hermanson GT (2013) Zero-length crosslinkers. In: Hermanson GT (ed) Bioconjugate techniques, 3rd edn. Elsevier, New York, pp 259–273
Landegren U, Nilsson M (1997) Locked on target: strategies for future gene diagnostics. Ann Med 29:585–590
Acknowledgements
This study was funded by the University of Illinois College of Medicine at Peoria start-up fund to PG, and a research grant from the Rising Tide Foundation (grant number CCR-17-400) to PG.
The authors would like to express their acknowledgement to Bryan Himmel from the University of Illinois at Urbana-Champaign, for language editing.
Author information
Authors and Affiliations
Contributions
Devised and designed the study: YS and PG. Performed the experiments: YS. Analyzed the images and data: YS. Drafted the manuscript: YS. Revised and agreed upon the manuscript: YS and PG. Provided reagents: PG.
Corresponding author
Ethics declarations
The authors declare that they have no competing interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 302 kb)
(MP4 38,713 kb)
Rights and permissions
About this article
Cite this article
Song, Y., Gyarmati, P. Visual detection of bacterial DNA using activated paper stripe. Microchim Acta 186, 642 (2019). https://doi.org/10.1007/s00604-019-3748-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3748-3