Abstract
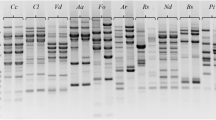
The ectomycorrhizal basidiomycete Tricholoma matsutake produces commercially valuable fruit bodies—matsutake—in Pinus sp. forest. Here we report that PCR with outward facing primers designed based on sequences comprising σ marY1 , the long terminal repeat of the gypsy-type retroelement marY1, specifies strains of T. matsutake. PCR with a primer based on the 22-bp sequence conserved at the 5′-end of σ marY1 conferred 73 reliable bands overall whose profiles depend upon strains of T. matsutake and T. magnivelare, the latter known as ‘American matsutake’. This PCR system gave no detectable band in any other species of Tricholoma tested, including T. bakamatsutake and T. fulvocastaneum, symbionts closely related to T. matsutake, as well as a host plant, Pinus densiflora. Similarly, PCR with a set of primers based on 26-bp and 28-bp sequences at bp 48–73 and bp 281–308 of σ marY1 , internal regions that are mutated in a variant of marY1, conferred 90 reliable bands only in strains of T. matsutake. Theoretically, PCR with the 22-bp primer would allow generation of 273, or 9.4×1021, types of polymorphism, and PCR with a combination of 26- and 28-bp primers, 290, or 1.2×1027 types. The probability of falsely specifying two different isolates as the same strain is <1/1021. While polymorphisms conferred by the primer based on the 5′ end of σ marY1 rather exhibit genetic conservation of a group of T. matsutake, those resulting from primers based on the internal sequences more clearly demonstrate intra-specific diversification. Both systems revealed that T. matsutake is divergent within the species. Ectomycorrhizas formed between P. densiflora and T. matsutake were identified by the PCR systems developed in the present study. This method, using σ marY1 as a genetic marker, is useful in analyzing the diversity of T. matsutake, monitoring the behavior of individual mycorrhizas, and specifying the ecological background of fruit bodies traded in markets.





Similar content being viewed by others
References
Babasaki K, Masuno K, Murata H (2003) Interactions of heterologous mycelia colonized in the substrate govern fruit body production in the cultivated homobasidiomycete Pholiota nameko. Biosci Biotechnol Biochem 67:100–106
Boeke JD (1989) Transposable elements in Saccharomyces cerevisiae. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington D.C., pp 335–374
Bushman F (2002) Lateral DNA transfer: mechanisms and consequences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 1–448
Díez J, Béguiristain T, Tacon FL, Casacuberta JM, Tagu D (2003) Identification of Ty1-copia retrotransposons in three ectomycorrhizal basidiomycetes: evolutionary relationships and use as molecular markers. Curr Genet 43:34–44
Dobinson KF, Harris RE, Hamer JE (1993) Grasshopper, a long terminal repeat (LTR) retroelement in the phytopathogenic fungus Magnaporthe grisea. Mol Plant Microbe Interact 6:114–126
Esser K, Blaich R (1994) Heterogenic incompatibility in fungi. In: Wessels JGH, Meinhardt F, Esser K, Lemke PA (eds) The Mycota I, a comprehensive treatise on fungi as experimental systems for basic and applied research: growth, differentiation and sexuality. Springer, Berlin Heidelberg New York, pp 211–232
Farman ML, Tosa Y, Nitta N, Leong SA (1996) MAGGY, a retrotransposon in the genome of the rice blast fungus Magnaporthe grisea. Mol Gen Genet 251:665–674
Fukuda M, Fukumasa-Nakai Y (1996) Genetic evidence supporting the multicellular origin of fruiting body formation in the basidiomycete Lentinula edodes. Mokuzai Gakkaishi 42:1025–1028
Guerin-Laguett A, Vaario LM, Gill WM, Lapeyrie F, Matsushita N, Suzuki K (2000) Rapid in vitro ectomycorrhizal infection on Pinus densiflora roots by Tricholoma matsutake. Mycoscience 41:389–393
Guerin-Laguett A, Matsushita N, Kikuchi K, Iwase K, Lapeyrie F, Suzuki K (2002) Identification of a prevalent Tricholoma matsutake ribotype in Japan by rDNA IGS1 spacer characterization. Mycol Res 106:435–443
Hamer JE (1991) Molecular probes for rice blast disease. Science 252:632–633
Hamer JE, Farrall L, Orbach MJ, Valent B, Chumley FG (1989) Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci USA 86:9981–9985
Hosford D, Pilz D, Molina R, Amaranthus M (1997) Ecology and management of the commercially harvested American matsutake mushroom. USDA-Forest service PNW-GTR412, pp 1–68
Kachroo P, Leong SA, Chattoo BB (1994) Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea. Mol Gen Genet 245:339–348
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kårén O, Högberg N, Dahlberg A, Jonsson L, Nylund J-E (1997) Inter- and intraspecific variation in the ITS region of rDNA of ectomycorrhizal fungi in Fennoscandia as detected by endonuclease analysis. New Phytol 136:313–325
Lagemaat LN van de, Landry JR, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Levy M, Romao J, Marchetti MA, Hamer JE (1991) DNA fingerprinting with a dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell 3:95–102
Lewin B (2004) Gene VIII. Pearson Prentice Hall/Pearson Education, Upper Saddle River, N.J., pp 1–1027
Lian C, Hogetsu T, Matsushita N, Guerin-Laguette A, Suzuki K, Yamada A (2003) Development of microsatellite markers from an ectomycorrhizal fungus, Tricholoma matsutake, by an ISSR-suppression-PCR method. Mycorrhiza 13:27–31
Murata H, Miyazaki Y (2001) Expression of marY1, a gypsy-type LTR-retroelement from the ectomycorrhizal homobasidiomycete Tricholoma matsutake, in the budding yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem 65:993–995
Murata H, Miyazaki Y (2004) σ marY1 , the LTR of the gypsy-type retroelement marY1 from the basidiomycete Tricholoma matsutake, allows multicopy DNA integration in Lentinula edodes. Biosci Biotechnol Biochem 68:218–221
Murata H, Yamada A (1999) Identification of ectomycorrhizae formed between Tricholoma matsutake and Pinus densiflora by polymerase chain reaction (PCR) targeting retroelement coding regions. Mycoscience 40:531–534
Murata H, Yamada A (2000) marY1, a member of the gypsy group of long terminal repeat retroelements from the ectomycorrhizal basidiomycete Tricholoma matsutake. Appl Environ Microbiol 66:3642–3645
Murata H, Yamada A, Babasaki K (1999) Identification of repetitive sequences containing motifs of retrotransposons in the ectomycorrhizal basidiomycete Tricholoma matsutake. Mycologia 91:766–775
Murata H, Miyazaki Y, Yamada A (2001) marY2N, a LINE-like non-long terminal repeat (non-LTR) retroelement from the ectomycorrhizal homobasidiomycete Tricholoma matsutake. Biosci Biotechnol Biochem 65:2301–2305
Nekrutenko A, Wen-Hsiung L (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17:619–621
Ogawa M (1975) Microbial ecology of mycorrhizal fungus-Tricholoma matsutake (Ito et Imai) Sing. in pine forest. II. Mycorrhiza formed by T. matsutake. Bull Gov For Exp Station 278:21–80
Ohta A (1994) Production of fruit-bodies of a mycorrhizal fungus, Lyophyllum shimeji, in pure culture. Mycoscience 35:147–151
Ohta A (1998) Fruit-body production of two ectomycorrhizal fungi in the genus Hebeloma in pure culture. Mycoscience 39:15–20
Peabody RB, Peabody DC, Sicard KM (2000) A genetic mosaic in the fruiting stage of Armillaria gallica. Fungal Genet Biol 29:72–80
Richardson DM (1998) Ecology and biogeography of Pinus. Cambridge University Press, Cambridge, pp 1–527
Shimamura M, Yasue H, Ohshima N (1997) Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature 388:666–670
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopathol 29:443-467
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res 18:6531–6535
Yamada A, Maeda K, Ohmasa M (1999) Ectomycorrhiza formation of Tricholoma matsutake isolates on seedlings of Pinus densiflora in vitro. Mycoscience 40:455–463
Yamada A, Kobayashi H, Murata H (2003) Tricholoma matsutake IFO6933 and IFO30604, ‘matsutake’ isolates that have been maintained on slants and widely used in vitro for a quarter to half a century, can form ectomycorrhizae in Pinus densiflora. Mycoscience 44:249–251
Acknowledgements
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fishery of Japan. The authors thank members of The Forest Experiment Station located in the following prefectures in Japan for the supply of fungal strains: Chiba, Fukui, Hiroshima, Hyogo, Iwate, Kyoto, Okayama, Shiga, Tokushima, Yamaguchi and Wakayama.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Murata, H., Babasaki, K. & Yamada, A. Highly polymorphic DNA markers to specify strains of the ectomycorrhizal basidiomycete Tricholoma matsutake based on σ marY1 , the long terminal repeat of gypsy-type retroelement marY1. Mycorrhiza 15, 179–186 (2005). https://doi.org/10.1007/s00572-004-0319-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-004-0319-0