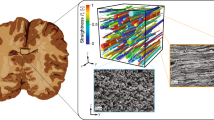
Abstract
A modular pipeline for improving the constitutive modelling of composite materials is proposed.The method is leveraged here for the development of subject-specific spatially-varying brain white matter mechanical properties. For this application, white matter microstructural information is extracted from diffusion magnetic resonance imaging (dMRI) scans, and used to generate hundreds of representative volume elements (RVEs) with randomly distributed fibre properties. By automatically running finite element analyses on these RVEs, stress-strain curves corresponding to multiple RVE-specific loading cases are produced. A mesoscopic constitutive model homogenising the RVEs’ behaviour is then calibrated for each RVE, producing a library of calibrated parameters against each set of RVE microstructural characteristics. Finally, a machine learning layer is implemented to predict the constitutive model parameters directly from any new microstructure. The results show that the methodology can predict calibrated mesoscopic material properties with high accuracy. More generally, the overall framework allows for the efficient simulation of the spatially-varying mechanical behaviour of composite materials when experimentally measured location-specific fibre geometrical characteristics are provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Composite materials, often consisting of inclusions or fibres embedded within a matrix with different mechanical properties, are notoriously difficult to model accurately with a single constitutive law. This challenge arises from the complex interactions between the different constituent materials and their respective orientations, generally leading to nonlinear, loading asymmetric, interdependent mechanical behaviours. Furthermore, in the case of anisotropic materials, e.g., with fibre inclusions, most of the established models assume uniform orientation within the structure of interest, setting aside the additional complexity of its underlying variability.
An extensively studied fibre-based composite material with high variability in orientation distribution is the brain white matter. A typical adult human brain contains an estimated \({\sim }\)100 billion neurons embedded within a matrix of glial cells [12]. A key component of neurons is the long, slender axon, which conducts electrical impulses along its length and is protected by a lipid-rich white substance called myelin which gives white matter its colour. The volume fraction of axons typically makes up for approximately 20% of the brain’s total volume. In comparison to axons, blood vessels only contribute about 3% to the brain’s total volume, and the remaining proportion primarily consists of glial cells [9, 12].
Although the availability of high-quality experimental data for the mechanical properties of white matter is limited due to the rapid change of its mechanical properties which occurs after removal from the brain [59], experiments consistently agree that it exhibits a nonlinear strain rate dependency and is close to incompressible [13] (although the latter is open for discussion when one accounts for the biphasic nature of the tissue). These key properties are captured well by hyperelastic models, with Ogden’s hyperelastic model being particularly prevalent in the literature and often used in both microscopic representative volume elements (RVEs) and mesoscopic constitutive models [6, 11, 18, 45, 48]. Much work has been done at both scales.
At the microscale, most of the proposed RVEs aim at capturing the region-dependent white matter microstructural features and focus on a select few features in order to reduce the complexity and number of dependent variables. Karami et al. [28] developed a repeating unit cell to investigate the effect of varying amounts of fibre undulation, which was later built upon by Pan et al. [44] by implementing partial coupling between the matrix and fibres rather than perfect bonding. This complex strain dependent coupling was also characterised by Bain et al. [4]. Fibre orientation has been shown to have a particularly significant contribution to stiffness, with higher degrees of anisotropy leading to higher stiffness [22]. However, not all microstructures have a significant effect on the mechanical response; Ho and Kleiven [24] included vasculature geometry from two angiographies in their model, but determined that it had a minimal effect on the dynamic response of the brain. Yousefsani et al. [56] modelled a histology-informed probabilistic fibre diameter distribution, and although this resulted in different stress concentrations, the RVE’s homogenised response was almost identical to the models using uniform diameter distributions with the same volume fraction. Although volume fraction estimates based on histology reports have been used, no studies were found which use directly and dynamically regional volume fraction estimates from dMRI scans. Even then, the unrealistically high computational cost of modelling a library of RVEs with all possible combination of axonal density, distribution and orientation means that mesoscopic models are generally more often used.
By virtue of the aimed homogenisation, current continuum mesoscopic constitutive models for white matter typically include a smaller variety of microstructure-based parameters. Chatelin et al. [6] developed a constitutive model which describes the matrix’ strain energy contribution via the Mooney–Rivlin hyperelastic formulation, and the fibres’ contributions via an exponential function of the mean fibre stretch, as proposed by Puso and Weiss [49]. However, the model inherently assumes an underlying uniaxial fibre distribution. Garcia-Gonzalez et al. [18] developed a model for transversely isotropic soft tissues using a similar approach, but with the popular Holzapfel–Gasser–Ogden model [13] to represent the fibres’ strain energy contributions. The matrix’ contribution was found to be best represented by Ogden’s hyperelastic formulation, when compared to other commonly-used hyperelastic models such as Mooney–Rivlin and Gent. This approach uses the generalised structure tensor (GST) method to allow information about fibre orientation distribution to be taken into account. The other common alternative to GST is angular integration. Angular integration involves the integration over the distribution of fibres at every iteration of the constitutive model. Although this provides a more accurate result, the required computational cost is significantly higher due to the integrals involved [27]. One overarching issue is that although almost all of the investigated constitutive models demonstrate a good fit with experimental data in one or two loading directions, there is significant disagreement when loading in tension, compression and shear in multiple directions [7]. This suggests that there is a lot of potential progress yet to be made towards more accurate constitutive models.
So as to ensure that the effects of mechanical loads on the spatially mechanically varying white matter are properly captured (e.g., impact in the context of traumatic brain injury, or pressure increase in the context of stroke), mechanical simulations making use of the finite element method (FEM) need to use accurate constitutive models of the brain at both macroscopic and microscopic levels. This multiscale link is particularly important here as the source of anisotropy is the component that confers the biological function, i.e., the axons. Neither the traditional RVE approach nor a general mesoscopic model are thus directly usable. One proposed way of achieving this relies on directly translating data obtained from magnetic resonance imaging (MRI) and diffusion MRI (dMRI) scans to a constitutive law, so that each element of a macroscopic FEM head model is governed by a slightly different constitutive law based on local axonal tract information. Each constitutive law then represents the microscopic structure and associated mechanical response of the tissue in each region, resulting in a heterogeneous anisotropic macroscopic white matter model. The inherent difficulty is to accurately predict how microstructural variations affect the local stresses without requiring a full set of RVE FEM simulations (for each loading case, e.g., tension, compression, shearing). One way to achieve this is to make use of machine learning.
The application of machine learning and big data in materials science and mechanics is gaining popularity, with modern computers able to process vast amounts of data in increasingly less time. Martín-Guerrero et al. [39] demonstrated the feasibility of predicting the deformation on human organs, given FEM data, information about the geometry, material properties, and loads applied, while Liang et al. [34] focussed on predicting stress distributions in a two-dimensional mesh. However, the extension of these methods to develop and improve existing constitutive models has only become a topic of research in the last decade, in the context of data-driven simulations. Indeed, in computational mechanics, finding the adequate constitutive models for a given often RVE often requires FEM simulations. As this process can be lengthy and computationally expensive, Li et al. [33] proposed to use a self-consistent clustering analyses method to quickly create a database where the inputs (strain) are linked to the outputs (stress). A deep neural network was then trained to give accurate prediction of the stress-state for any given strain-state, thus bypassing the need for additional FEM simulations. Liu et al. [35] suggested a two-stage approach for predicting the behavior of general heterogeneous materials under irreversible processes such as inelastic deformation with great accuracy. In the training stage (also called offline stage), the data are efficiently compressed using a k-means clustering algorithm. In the online stage, a self-consistent clustering analysis derived via the homogenisation of each cluster through the Lippmann–Schwinger integral equation allows an efficient prediction of the material’s mechanical behavior. Self-consistent clustering analysis have also been used to predict the overall mechanical response of polycrystalline materials in a finite strain framework [57]. This method has also been applied for multi-scale damage modelling where crack paths are predicted for elastoplastic strain softening materials [36]. Liu et al. [37] developed a transfer-learning strategy that trains a neural network to predict the stress state of a RVE with a given volume fraction and extend it to any volume fraction. This issue is not limited to computational mechanics, see, e.g., the work of Lu et al. [38] on the electrical behaviour of graphene/polymer nanocomposite. In this work, the so-called “FE\(^{2}\)” problem is too time-consuming and, the process is instead replaced by a faster approach (up to four order of magnitudes) consisting in training a neural network to predict the current density of the RVE for any given electric field. Le et al. [32] used neural networks to predict accurately the effective potential of nonlinear elastic materials for homogenisation problems. Their numerical studies have shown great efficiency even in high dimensional spaces (up to 10). Principal component analysis was also shown to reduce the dimension of the parameter space [55]. In this work, a neural network was trained on a database created from FEM simulations. The neural network predicted the principal stresses arising in the RVE submitted to principal stretches (only three components). Even if the objectivity of the material law was approximately enforced, this neural network could be trained in a cost-effective manner while still providing good estimate of the constitutive response for heterogeneous structures. With the aim of taking into account the immediate past history of loading in an effective way, Ghaboussi and Sidarta [21] used nested adaptive neural networks to model the path-dependent constitutive behavior of geomaterials. Recently, another paradigm was developed by Kirchdoerfer and Ortiz [30] that bypasses the empirical material modelling step that links stresses and strains. Instead, only experimental data points are assigned to integration points taking into account the constraint that conservation laws must be enforced. This approach is free of any constitutive model assumption but requires enough experimental data to map the stress-strain space accurately.
The proposed work aims at providing the level of microstructural detail arising from the use of RVEs while using mesoscopic constitutive models for the macroscopic head scale FEM simulations. The link between both is created through a machine learning layer leveraging a library of precalculated microscopic RVEs used as training data linking dMRI parameters and microscopic RVE stress-strain curves for a chosen set of loading scenarios. The parameters of a mesoscopic constitutive law are optimised for each RVE by fitting the stress-strain curves of the constitutive law to those output by the RVE analyses. The overall combination of library of precalculated cases and machine learning layer allows for the development and implementation of a pipeline for extracting microstructural information from a dMRI scan, translating it into a continuum constitutive model for use in patient-specific macroscopic head models, by predicting directly the mesoscopic model parameters, without the need for further RVE simulation. Figure 1 shows the key stages and their relationships. Components have been assigned a letter for ease of reference.
The pipeline is split into five processes (see code download link in Appendix), each listed below. Square brackets indicate which pipeline components each script corresponds to, while parentheses indicate approximate number of lines of code. A brief description is also included, and each process is discussed in more detail in the following.
-
1.
Sampling [A, B]: This process samples a diffusion tensor, extracts the microstructural parameters, calls DigimatGenerator with these inputs, and then repeats.
-
2.
DigimatGenerator [C]: This process calls Digimat-FE to create some geometry with a set of given input parameters, saves the Parasolid geometry files, and closes Digimat.
-
3.
DigimatToAbaqus [D, E, F]: Process that controls Abaqus’ kernel. It imports the geometry files, sets up nine analyses, executes them, post-processes the output data to extract the corresponding stress-strain curves, and then repeats.
-
4.
Optimisation [G, H]: Process that implements the constitutive model and then iteratively calibrates the optimal constitutive model parameters for each set of stress-strain curves.
-
5.
Learning [I, J]: This process performs a machine learning algorithm that maps diffusion tensor parameters directly onto constitutive model parameters.
Section 2 presents the relevant microstructural parameters to be extracted from dMRI. Section 3 presents the library of RVE simulations against which the mesoscopic constitutive model, presented in Sect. 4, is calibrated by the calibration layer presented in Sect. 5. Finally, the machine learning layer for future prediction of parameters is developed and its performance, evaluated in Sect. 6. Section 7 concludes this work.
2 dMRI microstructural parameters
Among the different dMRI techniques available, diffusion tensor imaging (DTI) has gained a lot of momentum as being particularly fitted to the identification of axonal orientation in the white matter. In this technique, key axonal orientation parameters can be extracted from the diffusion tensor \({\varvec{D}}\). In particular, the mean orientation of each voxel’s axon distribution \({\varvec{a}}_1\) is given by the eigenvector corresponding to its largest eigenvalue \(\lambda _1\). Another parameter is the mean diffusivity, \({\bar{\lambda }}\), equal to the mean of the eigenvalues of \({\varvec{D}}\) [50]:
The final DTI parameter of importance is the fractional anisotropy \({ FA }\):
\({{ FA }}\) is essentially a normalised measure of the eigenvalues’ variance, and therefore \({ FA } \in [0, 1]\), where a value of zero represents isotropic spherical diffusion, and a value of one represents uniaxial diffusion [50].
The neurite orientation dispersion and density imaging DTI (NODDI-DTI) model developed by Edwards et al. [10] uses the fact that by simplifying the NODDI model to not include cerebrospinal fluid, it is possible to achieve a one-to-one mapping between DTI parameters, \(FA\) and \({\bar{\lambda }}\), and the intra-cellular volume fraction \(v\) as estimated from NODDI:
where \(d\) is the intrinsic free diffusivity, and is fixed at \(d = 1.7 \times 10^{-3}\) \(\hbox {mm}^{2}\,\,\hbox {s}^{-1}\) [10].
The rationale behind Eq. (3) is that axons have a higher diffusivity than the glial matrix in which they are embedded, and therefore the mean diffusivity and the axonal volume fraction within a voxel are intrinsically linked. However, it is important to note that, rather than the mean diffusivity defined in Eq. (1), a heuristically corrected mean diffusivity \({\bar{\lambda }_h}\) is used instead, defined as follows:
where \(\delta _{ij}\) is the Kronecker delta function. This modification attempts to correct for diffusional kurtosis, based on the assumption that the distribution of diffusion displacement in brain tissue is actually more peaked than a Gaussian distribution, and that this is not properly captured by DTI due to its single-shell acquisition protocol. Substitution leads to Eq. (5), defining intra-cellular volume fraction as a function of parameters extracted from DTI data:
Therefore, with the eigenvectors and eigenvalues of the diffusion tensor, the following parameters can be extracted: \({\varvec{a}}_1\), \(FA\), \(v\) and \({\bar{\lambda }}\), which are mean orientation, fractional anisotropy, axon volume fraction and mean diffusivity, respectively.
Data from the UK Biobank, a group-averaged dataset as part of their ongoing project to scan 100,000 predominantly-healthy individuals, which includes averaged DTI parameters for each voxel in the registered scans, was used here [41]. For the data analysis of the specific parameters, a stratified training data has been performed [53]. These parameters include mean diffusivity, as well as the eigenvalues and eigenvectors (\({\bar{\lambda }}, \lambda _1, \lambda _2, \lambda _3, {\varvec{a}}_1, {\varvec{a}}_2,\) and \({\varvec{a}}_3\)). It is also useful to notice that for the FEM simulations of the microscopic RVEs, the orientation eigenvectors do not need to be sampled. Indeed, setting \({\varvec{a}}_1 = [1, 0, 0]^\text {T}\), \({\varvec{a}}_2 = [0, 1, 0]^\text {T}\), and \({\varvec{a}}_3 = [0, 0, 1]^\text {T}\) for all diffusion tensors reduces the dimensionality of the input parameter space. To obtain the mechanical response of a rotated voxel, the response of the unrotated voxel can then be transformed onto the correct frame of reference through appropriate rotation, see Sect. 4. The three eigenvalues and the mean diffusivity thus need to be sampled, although choosing any three of these parameters fixes the fourth. The Sampling process is achieved via the following steps:
-
1.
Produce a cumulative distribution function (CDF) for the mean diffusivity,
-
2.
Sample mean diffusivity using inverse transform sampling,
-
3.
Produce a joint probability density function (PDF) for two of the eigenvalues,
-
4.
Sample two eigenvalues from the PDF using rejection sampling,
-
5.
Calculate the final eigenvalue.
This approach has several advantages. Firstly, by using kernel density estimation to produce the CDF and PDF, the values in the dataset can effectively be interpolated, which would not be possible using direct sampling or histograms. Secondly, once the CDF and PDF have been produced, diffusion tensors can be sampled in approximately 0.05 seconds–a negligible amount of time compared to any other step in the pipeline. Finally, it allows variance to be re-introduced to the dataset, which would otherwise represent mean values. However, it is important to note that the underlying distributions are not necessarily Gaussian and identically distributed, so a large non-averaged dataset would ideally be needed. Here, we use dataset ID 9028, which is available as bmri_group_means.zip via download,Footnote 1 see Fig. 1-A. Voxels were randomly sampled using a combination of inverse transform sampling and rejection sampling. The final sampling of 232 voxels was shown to represent adequately the overall distribution. The ranges for the parameters of interest were 0.0485 to 0.8574 for FA, and 0.0133 to 0.8686 for the volume fraction, see Fig. 1-B.
3 RVE model
After obtaining a suitable sampling and extracting the key parameters describing the underlying microstructural features from the dMRI scan, FEM models of the RVE of each voxel were generated. Each RVE was loaded in tension, compression, and shear in multiple directions to produce a set of nine stress-strain curves.
The computation time of running FEM analyses is high, however to be able to exploit the machine learning layer, a significant number of analyses needs to be run. As the execution of these analyses takes up the majority of the processing time in the pipeline, the trade-off between computational cost and the accuracy of each FEM analysis has to be considered when making any design decision.
The toolkit Digimat-FE of the Digimat software [8] was used to generate RVEs of composites with a range of input parameters, such as inclusion shape, size, spacing, tortuosity, and orientation. Although Digimat also has the ability to mesh the RVEs and run several pre-defined FEM analyses, it was found that the complexity of the required geometry leads to extremely small elements near boundaries. Due to the need for more options to resolve these issues, Digimat-FE was only used for geometry generation, see Fig. 1-C. The geometry was then exported via a set of Parasolid files to Abaqus [1] (Fig. 1-D), where the scripting interface was used to set up, run, and post-process a fully automated and customisable set of FEM analyses, see Fig. 1-E.
3.1 Topology
One dMRI post-processing methodology is AxCaliber [3], aimed at extracting accurate axonal diameter distributions, but requiring up to 30 h to do so. Furthermore, diameter distributions have been shown to have a minimal effect on mechanical properties [56]. As such, all axon diameters were set to a mean value of two micrometres as a first approximation.
With regards to modelling the shape of axons, future work may wish to include tortuosity to allow for more accurate geometry and higher volume fractions to be achieved, as remaining gaps might be able to be filled by curved axons. However, tortuosity was assumed here to be negligible for the size of RVE considered and straight cylindrical inclusions were chosen due to their simplicity. Several conditions were also enforced, such as a minimum separation distance equal to 1% of each axon’s diameter. This ensured that no unrealistic mechanical behaviour occurred as a result of axons being connected to each other.
To include an orientation distribution, Digimat allows an orientation tensor to be input. By assuming a simple model of axons acting as rods with negligible diffusion perpendicular to their longitudinal axis, the orientation tensor and diffusion tensor can be considered equivalent. For axonal orientation in white matter, the von Mises-Fisher distribution (a Gaussian distribution mapped onto a sphere) is typically assumed, but other more peaked distributions such as the Watson distribution have been used in the literature [58]. Digimat uses a proprietary methodology for sampling inclusion orientations from a provided orientation tensor. By generating a large number of axons, it was found that the distribution closely resembles a von Mises-Fisher distribution, albeit with more peakedness when \(FA \approx 1\). However, analysis of UK Biobank’s dMRI data set showed that \(FA\) rarely approaches one, and therefore this was not deemed to be a significant issue. However, it does suggest that a bespoke geometry generation tool may be desirable in future work.
Voxels in dMRI scans are typically cubes with side lengths in the order of magnitude of one millimetre, whereas axon diameters are in the order of magnitude of one micrometre. By definition, RVEs have to be as small as possible while still capturing the material’s mechanical behaviour and underlying distribution of axons. To determine an appropriate RVE size and mesh density, FEM simulations were run with different sizes and levels of refinement, and the results were analysed by considering the trade-off between computational cost and accuracy. To ensure the validity of these results, a set of differently sized RVEs all including uniaxially aligned fibres of diameter two micrometres with a fixed volume fraction of 15% were produced. Tensile loading was arbitrarily applied perpendicular to the fibres. Figure 2 summarises the key findings regarding the convergence of mesh size and RVE size. Figure 2a shows that, for uniaxially aligned fibres, a mesh size of 0.8 \(\upmu {\hbox {m}}\) is sufficiently accurate, with less refined meshes leading to under-prediction of the stresses in this case. Figure 2b implies that RVE size itself does not have a significant effect on the result. However, this initial investigation did not take into account the fibre orientation distribution or run time.
It was noted that small RVEs have fewer axons, and therefore their distribution may not properly capture the distribution from which they are sampled. Digimat-FE samples axons using an orientation tensor, but also outputs the actual orientation tensor of the sampled axons. Comparison of the average root mean squared (RMS) error of each element of these two orientation tensors allowed the accuracy of the RVE generation process to be quantified for each sized RVE. 15 RVEs of each size were thus generated with a spherical orientation distribution, and volume fraction of 15%. The average RMS error was calculated for each RVE size, and FEM analyses were performed to find an approximate run time in each case. The results are shown in Fig. 3, with an upper limit set to one hour per set of nine analyses run on 32 CPU cores. This was chosen as a low but feasible rate to generate training data.
Figure 3 shows that, due to the number of degrees of freedom increasing with the cube of the RVE side length, and the simulated time period increasing with the RVE size, the total execution time quickly becomes unmanageably large. Secondly, the distribution of axons does not perfectly match the input distribution, even for relatively large RVEs. An RVE side length of 30 \(\upmu {\hbox {m}}\) was thus chosen as a compromise between run time and average RMS error. All parameters discussed here lead to RVEs of the form shown in Fig. 4.
3.2 Finite element framework
Abaqus’s scripting interface was used to automate the FEM stage with a single Python script, from importing the geometry, through to exporting the stress-strain curves, see Fig. 1-F. The flowchart in Fig. 5 shows the main steps within the DigimatToAbaqus process.
A material model often used in white matter RVEs to describe the nonlinear stress-strain behaviour observed is Ogden’s hyperelasticity formulation. In this model, the material’s strain energy density \(\psi \) (used here for both axons and the surrounding matter) is given by:
where \(\lambda _1\), \(\lambda _2\) and \(\lambda _3\) are the principal stretches and where N, \(\mu _p\) and \(\alpha _p\) for \(p\in [1,N]\) are material parameters. The nearly incompressible behaviour was modelled by imposing a Poisson’s ratio of 0.49. Values of \(N = 1\), \(\mu _1 = 290.82\) Pa, and \(\alpha _1 = 6.19\) have previously been identified as suitable parameters for axons [40]. It has also been reported that the stiffness of axonal fibres is approximately three times higher than that of the matrix, and therefore a value of \(\mu = 96.94\) Pa was chosen for the matrix [2, 56]. As \(N=1\), here and subsequently the subscripts are dropped. It must be emphasised that, while other (potentially more adequate) models do exist for white matter, this work is not concerned with the identification of the microscopic constitutive behaviour, but rather with the establishement of a flexible multiscale framework for any chosen microscopic model. As such, while acknoweldging its limitations, this model along with he proposed parameters for the axons and the matrix were chosen as a first approximation.
The dynamic explicit solver of Abaqus was used with 30% uniaxial (free lateral face) stretch boundary conditions for the tension and compression cases. To determine the required displacement fields for pure shear, it is noted that for an incompressible RVE, shear in the X-Y direction is equivalent to stretching the RVE by \(\lambda _{xy}\) lambdaxy in the direction 45 degrees from the X-axis. This deformation is also equivalent to rotating by -45 degrees about the Z-axis, stretching along the X-axis by \(\lambda _{xy}\), and reversing the initial rotation. Working in terms of \(\lambda _{xy}\) simplifies the mathematics significantly. Combining these transformations results in each point being transformed by the matrix \({\varvec{F}}_{xy}\), defined by:
Pardis et al. [46] demonstrated using simple geometric relationships that the shear strain can then be calculated as \(\gamma _{xy} = \text {tan}\left( \alpha \right) = \frac{1}{2}\left( \lambda ^2_{xy} - \lambda ^{-2}_{xy}\right) \). By using \({\varvec{F}}_{xy}\) to calculate the final position of each edge, and noting that the displacement of surface nodes vary linearly across each face, it is then simple to derive displacement fields for each node as a function of their position. Similar to the tensile and compressive analyses, the stretch \(\lambda _{xy}\) is varied using Abaqus’s smooth step function to avoid excessive distortion at the start of each analysis up to a final stretch of \(\lambda = 1.3\) in the direction of interest in each case. Finally, to avoid rounding errors associated with the RVE side lengths being in the order of 30 \(\upmu {\hbox {m}}\), and for consistency, units of grams, millimetres, seconds, pascals, micronewtons and joules were adopted in Abaqus. Double precision was also used for all simulations.
With the aim to automatically run thousands of simulations while avoiding failing simulations, traditional meshing techniques (even with a set of drastic meshing criteria) was shown to be unmanageable. The embedded element technique, an alternative to direct meshing, has previously been used to improve the mesh quality and success rate when meshing composite materials [45]. Rather than the direct meshing approach of subtracting the axon geometry from the matrix, the axons are instead superimposed on top of the matrix, then coupled to the matrix, and finally their stiffness is adjusted to account for the redundancy. Yousefsani et al. [56] demonstrated that, for Ogden hyperelastic materials, the excess strain energy can be removed by adjusting the axon stiffness as follows:
where \(\mu _f^*\) is the adjusted stiffness of the superimposed fibres, \(\mu _f\) is the fibre stiffness, and \(\mu _m\) is the matrix stiffness. The method was found to give accurate results, while avoiding mesh generation failing. Extremely small elements (thus leading to extremely small stable time increment) in the fibres were identified and the problematic fibres were automatically removed (see the DigimatToAbaqus process). The algorithm takes advantage of the fact that hexahedral meshes are less computationally costly than tetrahedral meshes, but fail to mesh complex geometry. Conversely, tetrahedral meshes can mesh all geometries, but may lead to very small elements. Therefore, by applying a hexahedral mesh, then a tetrahedral mesh on failed parts, and finally removing parts with excessively small elements, the algorithm is able to almost guarantee a successfully meshed RVE with a sensible run time. It was found that removing only the three most problematic fibres reduced the critical element size from approximately \(10^{-11}\)m to \(10^{-8}\)m, resulting in 1,000 times fewer iterations, and therefore 1,000 times shorter run times. However, this affected the underlying axon orientation distribution. Therefore, RVEs were automatically discarded if too many axons needed to be removed to achieve a sensible run time. Also, it was noted that removal of axons could affect the input parameters such as volume fraction and \(FA\), so the process was adjusted to re-calculate and save these parameters. Ultimately, a total of 232 RVEs were analysed under nine loading conditions, producing a total of 2,088 stress-strain curves.
4 Mesoscopic constitutive model
A key part of the pipeline is to find the parameters of a constitutive model which provide an optimal match with the stress-strain curves produced by running FEM analyses on a suitable RVE. This is essentially a curve-fitting problem, in which the constitutive model is chosen specifically for this purpose. By carefully selecting and matching the boundary conditions of the RVE FEM model with the ones of the mesoscopic constitutive model, the parameters of the latter were then calibrated, see Fig. 1-G.
The proposed constitutive model is schematised in Fig. 6. In this approach, the deformation gradient tensor, \({\varvec{F}}= \frac{\partial {\varvec{x}}}{\partial {\varvec{X}}}\), is decomposed into an isochoric component, \({\varvec{F}}_\text {iso} = J^{-1/3}{\varvec{F}}\), and a volume-changing component, \({\varvec{F}}_\text {vol} = J^{1/3}{\varvec{I}}\), where \(J = \text {det}\left( {\varvec{F}}\right) \). The isochoric component can then be split into its elastic and viscous constituents, \({\varvec{F}}_\text {e}\) and \({\varvec{F}}_\text {v}\), respectively:
As the studied problem does not involve viscosity, here and subsequently, the second equality is simplified to \({\varvec{F}}_\text {e}{\varvec{F}}_\text {vol}\).
4.1 Matrix branch
The Helmholtz free energy function per unit reference volume \(\Psi \) is decomposed into the summation of the equilibrium and non-equilibrium components, \(\Psi _\text {Eq}\) and \(\Psi _\text {NEq}\), both described by the Ogden hyperelastic function:
where \(\mu _\text {Eq}\), \(\mu _\text {NEq}\), \(\alpha _\text {Eq}\) and \(\alpha _\text {NEq}\) are material constants, while \(\lambda ^\text {e}_i\) and \(\lambda ^\text {iso}_i\) are the principal stretches. The first Piola-Kirchhoff stress tensors can then be derived using:
where \({\mathbb {P}} = {\mathbb {I}} - \frac{1}{3}{\varvec{F}}^{-T}\otimes {\varvec{F}}\), \({\mathbb {I}}\) is a fourth-order unity tensor, while \({\varvec{N}}^\text {e}_i\) and \({\varvec{N}}^\text {iso}_i\) are the eigenvector matrices of the deformation gradient tensors. Finally, the overall Piola-Kirchoff stress tensor is given via \({\varvec{P}}_\text {M} = {\varvec{P}}_\text {Eq} + {\varvec{P}}_\text {NEq}\), which then yields the matrix Cauchy stress \(\varvec{\sigma }_\text {M}\) through the usual transformation. Details of this derivation are left to Garcia-Gonzalez and Jérusalem [17], along with the updating of \({\varvec{F}}_v\) via the viscous flow rule.
4.2 Fibre branch
The dispersion parameter \(\xi \in [0, 1]\) is defined as a function of the fractional anisotropy \(FA\) by Wright et al. [54]:
This allows a generalised structure tensor \({\varvec{A}}\) to be defined as \({\varvec{A}} = \xi {\varvec{I}} + \left( 1 - 3\xi \right) {\varvec{a}}_1 \varvec{\otimes } {\varvec{a}}_1\), where \({\varvec{a}}_1\) is the normalised direction vector of the fibres. Next, the Helmholtz free energy function used by Garcia-Gonzalez et al. [18] leads to a Cauchy stress contribution of the fibres as follows:
where \(k_1\) and \(k_2\) are material parameters and \(I^*_\text {4F} = \text {tr}\left( {\varvec{F}}_\text {F}{\varvec{A}}{\varvec{F}}^T_\text {F}\right) \) is the isochoric fourth strain invariant, related to the mean fibre stretch by \({\bar{\lambda }}_F = \sqrt{I^*_\text {4F}}\), and \(J_\text {F} = \text {det}\left( {\varvec{F}}_\text {F}\right) \).
Finally, summation of the two derived Cauchy stress tensors results in the deformed contribution: \(\varvec{\sigma } = \varvec{\sigma _\text {M} + \sigma _\text {F}}\). This model was implemented in MATLAB.
5 Calibration layer
The goal of the calibration layer is to find, for each RVE, a set of material parameters allowing the mesoscopic constitutive model to match all stress-strain curves as accurately as possible. In particular, for each RVE, six loading modes (and stress-strain curves) were simultaneously calibrated: X-tension, Y-compression, Z-compression, X-Y-shear, X-Z-shear and Y-Z-shear (thus avoiding potential fibre buckling cases not considered here), see Fig. 1-G.
The stress contribution of the fibres depends on two material parameters, \(k_1\) and \(k_2\). To ensure a positive stress contribution from the fibres, the bounds \(k_1 \in [0, \infty )\) and \(k_2 \in [0, \infty )\) were enforced. Any remaining values were set to match the Ogden coefficients used in the RVE analyses, such as the Ogden coefficients for the matrix, which were \(\mu = 96.94\) Pa and \(\alpha = 6.19\).
As one cannot a priori exclude the fact that the optimisation problem is nonlinear and non-convex, a derivative-free optimisation method was used. Covariance Matrix Adaptation Evolution Strategy (CMA ES), a stochastic evolutionary algorithm [23] (available as a fully-customisable MATLAB package), was chosen. CMA ES is a very efficient algorithm in small scale epistatic problems [31, 42]. An appropriate cost function should represent the actual cost of an incorrect value in its real-world application, which in this case is the cost of the constitutive model either under-predicting or over-predicting the stresses when modelling white matter. Normalisation is necessary here because an error of 10 Pa is a more significant error on a curve with a maximum stress of 10 Pa than one with a maximum of 100 Pa. Similarly, squared distances should be used instead of absolute values because a single error of 50 Pa should be worse than an error of 1 Pa at 50 points, as it may indicate significant local damage. Finally, a small error across all nine stress-strain curves should be penalised less than a single poorly fit curve, due to the latter potentially leading to false stress concentrations and significant local damage. Therefore, each graph’s score should be squared before being summed to find the total score. Using all of the above requirements leads to the following cost function:
The function is the sum of squares for each individual graph, but with normalisation such that each FEM graph is effectively scaled to have a maximum value of one. For a given RVE, \(G_j\) and \(F_j\) refer to the \(j^{th}\) FEM stress-strain curve, and the corresponding mesoscopic constitutive model stress-strain curve, respectively. \(k\) refers to the \(k^{th}\) data point on each stress strain curve.
The optimisation script automatically generates a table of training data (Fig. 1-H) which links the input parameters extracted from dMRI data to the corresponding constitutive model parameters \(k_1\) and \(k_2\). The results generated for the 232 RVEs are summarised in Table 1. The data are saved in comma separated value (CSV) format so that it can be read easily by the machine learning script.
The performance of the calibration layer, i.e., how well the constitutive model reflects the material behaviour simulated by the microscopic model, is shown in Fig. 7 where the stress-strain curves from Sect. 3 are compared with the stress-strain curves obtained by minimising Eq. (16). Figure 7a–c shows, respectively, the results of the calibration layer for the RVE that exhibits the lowest, mean (one case with a mean cost was chosen arbitrarily) and highest cost (see Eq. (16)) from the 232 analysed RVEs.
6 Machine learning layer
The goal of the final machine learning step is to identify trends or relationships between the input and output parameters using regression-based supervised machine learning techniques. The selection of the algorithms is based on those with an expected good performance on a relatively small dataset (232 data points). This consideration suggests to discard deep learning or even neural network-based techniques [51]. The final set of candidate algorithms includes two regressors with regularisation (LASSO [52] and Ridge [26]) and five ensemble algorithms (two boosting methods: Ada Boost [14] and Gradient Boosting [15]; a general Bagging algorithm using random subspaces replacement sampling [25]; and two ensemble methods based on trees: Random Forests in their regression version [5] and a randomised version, known as “Extremely Randomised Trees” or “Extra Trees” [20]). Several of these algorithms have also been applied to other problems in material sciences with small datasets [60]. In this study, all the algorithms are based on Python’s scikit-learn library implementation. The parameters used for these algorithms were the defaults ones, see Table 2.
To determine a suitable model, stratified repeated \(10\times 5\)-fold cross validation.Footnote 2 The average of the \(10 \times 5\) repetitions for a dataset of 232 data points minimises the variance, and keeps a reasonable number of training samples and enough validation data to control the bias [29]. The accuracy measure applied to this analysis is the coefficient of determination \(R^2\), whose score 1 indicates that the regression fits perfectly to the data and 0 or even negative values that the regression provides low quality fits.
Table 3 shows the mean \(R^2\) values for each of the target parameters plus and minus two standard deviations, indicating the 95% confidence intervals. It shows that both \(k_1\) and \(k_2\) were predicted reasonably well by all of the ensemble algorithms. The Gradient Boosting Regression algorithm using all of the inputs had the best result, and therefore this algorithm was chosen. These results are objectively very accurate for the amount of available data points and the number of input variables (5). The number of variables limits the effectiveness of the regularisation performed by both LASSO and Ridge, making them perform as regular linear regressors. On the contrary, those methods including repetitive regressors perform better and are more stable, in particular those with more regression components, such as the two boosting methods.
It is also worth emphasising that two of the inputs depend directly on the values of the three others (v, FA and the \(\lambda _i\)). In fact, the inclusion of a moderate number of redundant or derived features as an input for a machine learning algorithm is a well-known strategy to improve the quality of supervised methods, in particular, for those methods embedding regularisation or other feature selection mechanisms [47], which may identify the subset of these features that provides better classification/regression accuracy.
To visualise the Gradient Boosting Regressor algorithm’s output, the model was trained on the entirety of the training set, and values of the test set were predicted. Figure 8 shows a scatter plot of the test set, with the overlayed line showing the predicted values of \(k_1\). Although the model initially appears to overfit the data, a more in-depth look at the data indicates that the fluctuations on the prediction line are mostly due to different volume fractions.
7 Discussion
A proof-of-concept pipeline to improve constitutive models using automated FEM analyses, optimisation, and machine learning has been proposed, developed and implemented. By applying the pipeline to a natural composite material, brain white matter, stress-strain data have been generated to create a library of hundreds of RVEs. This library was found to provide sufficient training data to predict with a 96% accuracy material parameters for a mesoscopic constitutive model without the need for further RVE calculations. The approach was applied in this work to brain white matter through dMRI acquisition. Many recent computational head models assume isotropy (e.g., ref. [16]), or constant anisotropic propertiesFootnote 3 (e.g., ref. [19]), and all of them rely on a few experimental tests in a limited set of regions, while ignoring or simplifying the influence of axonal density in the locations of interest. These models thus involve a “leap of faith” when used for the entire white matter region. The proposed approach not only avoids calibrating the mechanical properties from microscopic model upscaling for all relevant microstructural configurations but also ensures its applicability to the entire brain in a timely and cost-effective manner.
Outside of the context of the brain, the proposed framework can be used for any composites material subject to microstructural variability, whether by design or through manufacturing uncertainty. In both cases, an experimental evaluation of the microstructural variations is enough to obtain, after RVE training data have been generated, an on-the-fly model adaptation when simulating meso- to macroscopic components.
A few limitations have been identified. A constant fibre diameter was used due to meshing difficulties, albeit comforted by literature findings (in the case of white matter) indicating a limited influence of diameter distribution on the mechanical response. In hindsight, however, this inherently limited the achievable volume fractions, as gaps were naturally left in the RVEs. This led to bias in the training data, with an unnaturally high proportion of RVEs with a volume fraction of around \(0.2\). As a result, histology-informed diameter distributions (or any microstructural counterpart for other composite materials) should be used whenever possible. In the particular case of brain white matter, fibre mechanical properties might also differ depending on the species or the brain region, and both matrix and fibres mechanical properties might actually be different for different measured diffusion tensors. While this sits outside of the scope of this work, the incorporation of these specificities in the proposed framework should be straightforward when available. Also, in reality, fibres can a priori buckle under compression in their longitudinal direction, and therefore have a negligible stress contribution relative to tensile longitudinal loading. To this end, the possibility of using different strain energy functions was researched, but a solution to properly capture this behaviour using a small RVE was ultimately not found. It is hypothesised, however, that this would not be an issue for stiff-fibre composite materials due to their higher buckling strength.
Data availability
The full code is available under academic license on http://jerugroup.eng.ox.ac.uk/?page_id=2563 or on the Oxford University Innovation Click-to-licence Portal https://process.innovation.ox.ac.uk/software/p/17979a/machine-learning-based-finite-element-constitutive-model-calibration-tool-for-composite-materials-(academic)/1.
Notes
A validation procedure in which the original dataset is divided into K folds (subsets) by stratified random sampling (ensuring that the original classes are proportionally represented in the same ratios that they appear in the original dataset) in each of the K subsets. Then, iteratively, K-1 folds are used to train the model and the remaining fold is used to validate. The process is repeated K times taking a different fold at a time for validation. The whole process of creating the K folds is repeated N times. The finals results are averaged from N \(\times \) K repetitions.
Only driven by the mean axonal tract orientation.
References
Abaqus (2013) Abaqus documentation. http://dsk.ippt.pan.pl/docs/abaqus/v6.13/books/gsk/ch10s06.html
Arbogast KB, Margulies SS (1999) A fiber-reinforced composite model of the viscoelastic behavior of the brainstem in shear. J Biomech 32:865–870
Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ (2008) AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Resonan Med 59:1347–1354
Bain AC, Shreiber DI, Meaney DF (2003) Modeling of microstructural kinematics during simple elongation of central nervous system tissue. J Biomech Eng 125:798–804
Breiman L (2001) Random forests. Mach Learn 45:5–32
Chatelin S, Deck C, Willinger R (2013) An anisotropic viscous hyperelastic constitutive law for brain material finite-element modeling. J Biorheol 27:26–37
de Rooij R, Kuhl E (2016) Constitutive modeling of brain tissue: current perspectives. Appl Mech Rev 68:010801
Digimat (2018) e-xstream engineering. https://www.e-xstream.com/products/digimat/about-digimat
Duval T, Stikov N, Cohen-Adad J (2016) Modeling white matter microstructure. Funct Neurol 31:217–228
Edwards LJ, Pine KJ, Ellerbrock I, Weiskopf N, Mohammadi S (2017) NODDI-DTI: estimating neurite orientation and dispersion parameters from a diffusion tensor in healthy white matter. Front Neurosci 11:720
El Sayed T, Mota A, Fraternali F, Ortiz M (2008) Biomechanics of traumatic brain injury. Comput Methods Appl Mech Eng 197:4692–4701
Filley C (2012) White matter structure and function. In: Filley C (ed) The behavioral neurology of white matter. Oxford University Press, Oxford, pp 23–41
Franceschini G, Bigoni D, Regitnig P, Holzapfel GA (2006) Brain tissue deforms similarly to filled elastomers and follows consolidation theory. J Mech Phys Solids 54:2592–2620
Freund Y, Schapire RE (1995) A decision-theoretic generalization of on-line learning and an application to boosting. In: European conference on computational learning theory. Springer, pp 23–37
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 2001:1189–1232
Garcia-Gonzalez D, Jayamohan J, Sotiropoulos S, Yoon S-H, Cook J, Siviour C, Arias A, Jérusalem A (2017) On the mechanical behaviour of peek and ha cranial implants under impact loading. J Mech Behav Biomed Mater 69:342–354. https://doi.org/10.1016/j.jmbbm.2017.01.012
Garcia-Gonzalez D, Jérusalem A (2019) Energy based mechano-electrophysiological model of CNS damage at the tissue scale. J Mech Phys Solids 125:22–37
Garcia-Gonzalez D, Jérusalem A, Garzon-Hernandez S, Zaera R, Arias A (2018a) A continuum mechanics constitutive framework for transverse isotropic soft tissues. J Mech Phys Solids 112:209–224
Garcia-Gonzalez D, Race NS, Voets NL, Jenkins DR, Sotiropoulos SN, Acosta G, Cruz-Haces M, Tang J, Shi R, Jérusalem A (2018b) Cognition based bTBI mechanistic criteria; a tool for preventive and therapeutic innovations. Sci Rep 8:10273
Geurts P, Ernst D, Wehenkel L (2006) Extremely randomized trees. Mach Learn 63:3–42
Ghaboussi J, Sidarta D (1998) New nested adaptive neural networks (nann) for constitutive modeling. Comput Geotech 22:29–52. https://doi.org/10.1016/S0266-352X(97)00034-7
Giordano C, Kleiven S (2014) Connecting fractional anisotropy from medical images with mechanical anisotropy of a hyperviscoelastic fibre-reinforced constitutive model for brain tissue. J R Soc Interface 11:20130914
Hansen N (2008) Covariance matrix adaptation - evolution strategy: Source code. https://www.lri.fr/hansen/cmaes_inmatlab.html
Ho J, Kleiven S (2007) Dynamic response of the brain with vasculature: a three-dimensional computational study. J Biomech 40:3006–3012
Ho TK (1998) The random subspace method for constructing decision forests. IEEE Trans Pattern Anal Mach Intell 20:832–844
Hoerl AE, Kennard RW (2000) Ridge regression: biased estimation for nonorthogonal problems. Technometrics 42:80–86
Holzapfel GA, Ogden RW (2017) On fiber dispersion models: exclusion of compressed fibers and spurious model comparisons. J Elast 129:49–68
Karami G, Grundman N, Abolfathi N, Naik A, Ziejewski M (2009) A micromechanical hyperelastic modeling of brain white matter under large deformation. J Mech Behav Biomed Mater 2:243–254
Kim J-H (2009) Estimating classification error rate: repeated cross-validation, repeated hold-out and bootstrap. Comput Stat Data Anal 53:3735–3745
Kirchdoerfer T, Ortiz M (2015) Data driven computational mechanics. Comput Methods Appl Mech Eng 304:81–101. https://doi.org/10.1016/j.cma.2016.02.001
LaTorre A, Muelas S, & Peña J M (2010) Benchmarking a mos-based algorithm on the bbob-2010 noiseless function testbed. In: Proceedings of the 12th annual conference companion on Genetic and evolutionary computation, pp 1649–1656
Le BA, Yvonnet J, He Q (2015) Computational homogenization of nonlinear elastic materials using neural networks. Int J Numer Methods Eng 104:1061–1084. https://doi.org/10.1002/nme.4953
Li H, Kafka O, Gao J et al (2019) Clustering discretization methods for generation of material performance databases in machine learning and design optimization. Comput Mech 64:281–305. https://doi.org/10.1007/s00466-019-01716-0
Liang L, Liu M, Martin C, Sun W (2018) A deep learning approach to estimate stress distribution: a fast and accurate surrogate of finite-element analysis. J R Soc Interface 15:20170844
Liu Z, Bessa M, Liu W (2016) Self-consistent clustering analysis: an efficient multi-scale scheme for inelastic heterogeneous materials. Comput Methods Appl Mech Eng 306:319–341. https://doi.org/10.1016/j.cma.2016.04.004
Liu Z, Fleming M, Liu W (2018) Computer methods in applied mechanics and engineering microstructural material database for self-consistent clustering analysis of elastoplastic strain softening materials. Comput Methods Appl Mech Eng 330:547–577. https://doi.org/10.1016/j.cma.2017.11.005
Liu Z, Wu C, Koishi M (2019) Transfer learning of deep material network for seamless structure-property predictions. Comput Mech 64:451–465. https://doi.org/10.1007/s00466-019-01704-4
Lu X, Giovanis D, Yvonnet J et al (2018) A data-driven computational homogenization method based on neural networks for the nonlinear anisotropic electrical response of graphene/polymer nanocomposites. Comput Mech 64:307–321. https://doi.org/10.1007/s00466-018-1643-0
Martín-Guerrero JD, Rupérez-Moreno MJ, Martinez-Martínez F, Lorente-Garrido D, Serrano-López AJ, Monserrat C, Martínez-Sanchis S, Martínez-Sober M (2016) Machine learning for modeling the biomechanical behavior of human soft tissue. In: 2016 IEEE 16th international conference on data mining workshops (ICDMW), pp 247–253
Meaney DF (2003) Relationship between structural modeling and hyperelastic material behavior: application to CNS white matter. Biomech Model Mechanobiol 1:279–293
Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JLR, Griffanti L, Douaud G, Okell TW, Weale P, Dragonu I, Garratt S, Hudson S, Collins R, Jenkinson M, Matthews PM, Smith SM (2016) Multimodal population brain imaging in the UK biobank prospective epidemiological study. Nat Neurosci 19:1523–1536
Muelas S, La Torre A, & Peña J-M (2009) A memetic differential evolution algorithm for continuous optimization. In: 2009 Ninth international conference on intelligent systems design and applications. IEEE, pp 1080–1084
Narayanan RT, Udvary D, Oberlaender M (2017) Cell type-specific structural organization of the six layers in rat barrel cortex. Front Neuroanat 11:91
Pan Y, Shreiber DI, Pelegri AA (2011) A transition model for finite element simulation of kinematics of central nervous system white matter. IEEE Trans Bio-med Eng 58:3443–3446
Pan Y, Sullivan D, Shreiber DI, Pelegri AA (2013) Finite element modeling of CNS white matter kinematics: use of a 3D RVE to determine material properties. Front Bioeng Biotechnol 1:19
Pardis N, Ebrahimi R, Kim HS (2017) Equivalent strain at large shear deformation: theoretical, numerical and finite element analysis. J Appl Res Technol 15:442–448
Parr R, Painter-Wakefield C, Li L, & Littman M (2007) Analyzing feature generation for value-function approximation. In: Proceedings of the 24th international conference on Machine learning, pp 737–744
Prange MT, Margulies SS (2002) Regional, directional, and age-dependent properties of the brain undergoing large deformation. J Biomech Eng 124:244–252
Puso MA, Weiss JA (1998) Finite element implementation of anisotropic quasi-linear viscoelasticity using a discrete spectrum approximation. J Biomech Eng 120:62–70
Soares JM, Marques P, Alves V, Sousa N (2013) A hitchhiker’s guide to diffusion tensor imaging. Front Neurosci 7:31
Sun C, Shrivastava A, Singh S, Gupta A (2017) Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE international conference on computer vision, pp 843–852
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J Roy Stat Soc: Ser B (Methodol) 58:267–288
Witten IH, Frank E (2005) Data mining: practical machine learning tools and techniques, 2nd edn. Morgan Kaufmann, Burlington
Wright RM, Post A, Hoshizaki B, Ramesh KT (2013) A multiscale computational approach to estimating axonal damage under inertial loading of the head. J Neurotrauma 30:102–118
Yang H, Guo X, Tang S, Liu W (2019) Derivation of heterogeneous material laws via data-driven principal component expansions. Comput Mech 104:365–379. https://doi.org/10.1007/s00466-019-01728-w
Yousefsani SA, Shamloo A, Farahmand F (2018) Micromechanics of brain white matter tissue: a fiber-reinforced hyperelastic model using embedded element technique. J Mech Behav Biomed Mater 80:194–202
Yu C, Kafka O, Liu W (2019) Self-consistent clustering analysis for multiscale modeling at finite strains. Comput Methods Appl Mech Eng 349:339–359. https://doi.org/10.1016/j.cma.2019.02.027
Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC (2012) NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 61:1000–1016
Zhang W, Liu Y-F, Liu L-F, Niu Y, Ma J-L, Wu C-W (2017) Effect of vitro preservation on mechanical properties of brain tissue. J Phys Conf Ser 842:012005
Zhang Y, Ling C (2018) A strategy to apply machine learning to small datasets in materials science. NPJ Comput Mater 4:1–8
Acknowledgements
The authors would like to thank Dr. Natalie Voets for the fruitful discussion on dMRI, and Dr. Daniel Garcia-Gonzalez for the discussions on the constitutive model.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Field, D., Ammouche, Y., Peña, JM. et al. Machine learning based multiscale calibration of mesoscopic constitutive models for composite materials: application to brain white matter. Comput Mech 67, 1629–1643 (2021). https://doi.org/10.1007/s00466-021-02009-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00466-021-02009-1