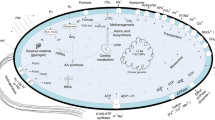
Abstract
Clostridium butyricum can convert glycerol into 1,3-propanediol, thereby generating unfortunately a high amount of acetate, formate and butyrate as inhibiting by-products. We have proposed a novel mixed culture comprising C. butyricum and a methane bacterium, Methanosarcina mazei, to relieve the inhibition and to utilise the by-products for energy production. In order to examine the efficiency of such a mixed culture, metabolic modelling of the culture system was performed in this work. The metabolic networks for the organisms were reconstructed from genomic and physiological data. Several scenarios were analysed to examine the preference of M. mazei in scavenging acetate and formate under conditions of different substrate availability, including methanol as a co-substrate, since it may exist in glycerol solution from biodiesel production. The calculations revealed that if methanol is present, the methane production can increase by 130%. M. mazei can scavenge over 70% of the acetate secreted by C. butyricum.










Similar content being viewed by others
References
Ashworth W (1991) The encyclopaedia of environmental studies. Facts on File, New York
Calow P (1998) The encyclopaedia of ecology and environmental management. Oxford Blackwell Science, Oxford
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280
Schink B (2002) Synergistic interactions in the microbial world. Antonie van Leeuvenhoek 81:257–261
Stouthamer AH (1988) Bioenergetics and yields with electron acceptors other than oxygen. In: Erickson LE, Fung DYC (eds) Handbook on anaerobic fermentations. Marcel Dekker, New York
Misoph M, Drake HL (1996) Effect of CO2 on the fermentation capacities of the acetogen Peptostreptococcus productus U-1. J Bacteriol 178:3140–3145
Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl D (2007) Metabolic modeling of a mutualistic community. Mol Syst Biol 3:92
Kim BH, Gadd GM (2008) Bacterial physiology and metabolism. Cambridge University Press, New York
Biebl H, Marten S, Hippe H, Deckwer W-D (1992) Glycerol conversion to 1, 3-propanediol by newly isolated clostridia. Appl Microbiol Biotechnol 36:592–597
Zeng A-P, Ross A, Biebl H, Tag C, Guenzel B, Deckwer W-D (1994) Multiple product inhibition and growth modeling of Clostridium butyricum and Klebsiella pneumoniae in glycerol fermentation. Biotechnol Bioeng 44:902–911
Nishio NK, Kuroda K, Nagai S (1990) Methanogenesis of Glucose by defined thermophilic coculture of Clostridium thermoaceticum and Methanosarcina sp. J Ferm Bioeng 70:398–403
Weimer PJ, Zeikus JG (1977) Fermentation of cellulose and cellobiose by Clostridium thermocellum in absence and presence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol 33:289–297
Yang ST, Tang IC (1990) Methanogenesis from lactate by a co-culture of Clostridium formicoaceticum and Methanosarcina mazei. Appl Microbiol Biotechnol 35:119–123
Yang Y, Tsukahara K, Sawayama S (2008) Biodegradation and methane production from glycerol-containing synthetic wastes with fixed-bed bioreactor under mesophilic and thermophilic conditions. Proc Biochem 43:362–367
Zeng A-P (1996) Pathway and kinetic analysis of 1, 3-propanediol production from glycerol fermentation by Clostridium butyricum. Bioprocess Eng 14:169–175
Jung K (2001) Quantitative Physiologie und metabolische Stofflußmodellierung der mikrobiellen Herstellung von 1,3-propandiol. PhD thesis, Gesselschaft für Biotechnologische Forschung mbH in Braunschweig
Zhang Q, Teng H, Sun Y, Xiu Z, Zeng A-P (2008) Metabolic flux and robustness analysis of glycerol metabolism in Klebsiella pneumoniae. Bioprocess Biosyst Eng 31:127–135
Neidhardt FC, Ingraham JL, Schaechter M (1990) Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland
Deppenheimer U (2003) Redox-driven proton translocation in methanogenic Archea. Cell Mol Life Sci 59:1513–1533
Feist AM, Scholten JCM, Pallsson BO, Brockman FJ, Ideker T (2006) Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 1:4
Ahrens K, Menzel K, Zeng A-P, Deckwer W-D (1998) Kinetic, dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture: III. Enzymes and fluxes of glycerol dissimilation and 1,3-propanediol formation. Biotechnol Bioeng 59:544–552
Hartlep M (2006) Prozessenwicklung und metabolic engineering der mikrobiellen Produktion von 1,3-Propandiol. PhD thesis, Technische Universität Carolo-Wilhelmina zu Braunschweig
Zeng A-P (1995) Effect of CO2 absorption on the measurement of CO2 evolution rate in aerobic and anaerobic continuous cultures. Appl Microbiol Biotechnol 42:688–691
Solomon BO, Zeng A-P, Biebl H, Schlieker H, Posten C, Deckwer W-D (1995) Comparison of the energetic efficiencies of hydrogen and oxychemicals formation in Klebsiella pneumoniae and Clostridium butyricum during anaerobic growth on glycerol. J Biotechnol 39:107–117
Rajoka MI, Tabassum R, Malik KA (1999) Enhanced rate of methanol and acetate uptake for production of methane in batch cultures using Methanosarcina mazei. Bioresour Technol 67:305–311
Weimer PJ, Zeikus JG (1978) One carbon metabolism in methanogenic bacteria. Cellular characterization and growth of Methanosarcina barkeri. Arch Microbiol 119:49–57
Schauer NL, Ferry JG (1980) Metabolism of formate in Methanobacterium formicicum. J Bacteriol 142:800–807
Acknowledgments
Marcin Bizukojc wishes to express his gratitude to Deutscher Akademischer Austausch Dienst (DAAD) for the financial support during his stay at the Hamburg University of Technology (special scholarship programme “Modern Applications of Biotechnology” PKZ no. A/07/97472). This work was also supported by the German Research Foundation (DFG project no. ZE 542/2-1) and the European 7 Framework Research Programme (project no. 212671-Propanergy).
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Appendices
Abbreviations used in the graphs, tables and text (for protein amino acids, standard abbreviations were used)
- 3-HPA:
-
3-Hydroxypropionaldehyde
- 3PG:
-
3-Phosphoglycerate
- AcCoA, CH3COCoA:
-
Acetyl-CoA
- AKG:
-
α-Ketoglutarate
- Asa:
-
l-Aspartate 4-semialdehyde
- CH2H4MPT:
-
Methylenetetrahydromethanopterin
- CH3CoM:
-
Methyl coenzyme M
- CH3H4MPT:
-
Methyltetrahydromethanopterin
- CH3H4SPT:
-
Methyltetrahydrosarcinopterin
- CHR:
-
Chorismate
- CO:
-
Carbon monoxide
- DHA:
-
Dihydroxyacetone
- DHAP:
-
Dihydroxyacetonephosphate
- E4P:
-
Erythrose-4-phosphate
- EtOH:
-
Ethanol
- F420(H), F420(red):
-
Coenzyme F420 (reduced)
- F6P:
-
Fructose-6-phosphate
- FBP:
-
Fructose-1,6-biphosphate
- Fd(H), FdH:
-
Ferredoxin (reduced)
- FORM:
-
Formic acid (formate)
- FUM:
-
Fumarate
- G6P:
-
Glucose-6-phosphate
- H4MPT:
-
Tetrahydromethanopterin
- H4SPT:
-
Tetrahydrosarcinopterin
- HAc:
-
Acetic acid (acetate)
- HBu:
-
Butyric acid (butyrate)
- HCH4MPT:
-
Methenyltetrahydromethanopterin
- HCOH4MPT:
-
Formyltetrahydromethanopterin
- Hse:
-
Homoserine
- ICT:
-
Isocitrate
- Ind:
-
Indole
- kiV:
-
α-Ketoisovalerate
- LAC:
-
Lactic acid (lactate)
- MeOH:
-
Methanol
- MethPhen(H), dHMePhe:
-
Methanophenazine (reduced)
- OAA:
-
Oxalacetate
- PEP:
-
Phosphoenolpyruvate
- PPA:
-
Prephenate
- PRPP:
-
Phosphoribosyl pyrophosphate
- PYR:
-
Pyruvate
- qCH3COOH:
-
Specific acetate uptake rate
- qCH3OH:
-
Specific methanol uptake rate
- qCH4 :
-
Specific methane production rate
- qCO2 :
-
Specific carbon dioxide production/uptake rate
- qH2 :
-
Specific hydrogen uptake rate
- qHCOOH:
-
Specific formate uptake rate
- RIB5P:
-
Ribose-5-phosphate
- S7P:
-
Sedoheptulose-7-phosphate
- SKA:
-
Shikimate
- SUC-CoA:
-
Succinyl-CoA
- X5P:
-
Xylose-5-phosphate
- \( {\text{Y}}_{{{\text{CH}}_{ 4} / {\text{CH}}_{ 3} {\text{COOH}}}} \) :
-
Methane to acetate yield coefficient
- \( {\text{Y}}_{{{\text{CH}}_{ 4} / {\text{CH}}_{ 3} {\text{OH}}}} \) :
-
Methane to methanol yield coefficient
- \( {\text{Y}}_{{{\text{CH}}_{ 4} / {\text{CO}}_{ 2} }} \) :
-
Methane to carbon dioxide yield coefficient
- \( {\text{Y}}_{{{\text{CH}}_{ 4} / {\text{HCOOH}}}} \) :
-
Methane to formate yield coefficient
- \( {\text{Y}}_{{{\text{CH}}_{ 4} / {\text{X}}}} \) :
-
Methane to biomass yield coefficient
- \( {\text{Y}}_{{{\text{CO}}_{ 2} / {\text{H}}_{ 2} }} \) :
-
Carbon dioxide to hydrogen yield coefficient
- \( {\text{Y}}_{{{\text{CO}}_{ 2} / {\text{HCOOH}}}} \) :
-
Carbon dioxide to formate yield coefficient
- μMM :
-
M. mazei specific biomass growth rate
Abbreviations used in the formulation of metabolic network only (in the stoichiometric equations): exchanged metabolites
The notation was so designed that the names of all metabolites, which belong to C. butyricum start with letter “P” and so do the names of reaction rates listed down in Tables 1 and 2. For M. mazei, it is the letter “M”.
- CO2(exch):
-
Carbon dioxide (the exchanged pool)
- FORM(exch):
-
Formate (the exchanged pool)
- H2(exch):
-
Hydrogen (the exchanged pool)
- HAc(exch):
-
Acetate (the exchanged pool)
Intracellular metabolites of M. mazei
- M_3PG:
-
3-Phosphoglycerate
- M_AcCoA:
-
Acetyl coenzyme A
- M_AcP:
-
Acetophosphate
- M_aIVA:
-
α-Ketovalerate
- M_AKG:
-
Aconitate
- M_Ala:
-
Alanine
- M_Arg:
-
Arginine
- M_ASA:
-
l-Aspartate 4-semialdehyde
- M_Asn:
-
Asparagine
- M_Asp:
-
Aspartate
- M_ATP:
-
Adenosinetriphosphate
- M_CH3CoM:
-
Methyl coenzyme M
- M_CH3H4MPT:
-
Methyltetrahydromethanopterin
- M_CH3H4SPT:
-
Methyltetrahydrosarcinopterin
- M_CH3OH:
-
Methanol
- M_CH4 :
-
Methane
- M_CHR:
-
Chorismate
- M_CO:
-
Carbon monoxide
- M_CO2 :
-
Carbon dioxide
- M_Cys:
-
Cysteine
- M_dHMethphen:
-
Methanophenazine (reduced)
- M_DNA:
-
DNA
- M_E4P:
-
Erythrose-4-phosphate
- M_F420red:
-
Coenzyme F420 (reduced)
- M_F6P:
-
Fructose-6-phosphate
- M_FBP:
-
Fructose-1,6-diphosphate
- M_FdH:
-
Ferredoxin (reduced)
- M_FORM:
-
Formate
- M_FORMH4MPT:
-
Formyltetrahydromethanopterin
- M_FUM:
-
Fumarate
- M_G1P:
-
Glucose
- M_G6P:
-
Glucose-6-phosphate
- M_GA3P:
-
Glyceraldehyde phosphate
- M_Gln:
-
Glutamine
- M_Glu:
-
Glutamate
- M_Gly:
-
Glycine
- M_gly:
-
Glycogen
- M_H+ :
-
Hydrogen ions
- M_H2 :
-
Hydrogen
- M_HAc:
-
Acetic acid
- M_His:
-
Histidine
- M_Hse:
-
Homoserine
- M_Ile:
-
Isoleucine
- M_IND:
-
Indole
- M_Leu:
-
Leucine
- M_Lys:
-
Lysine
- M_MeH4MPT:
-
Methylenetetrahydromethanopterin
- M_Met:
-
Methionine
- M_MthH4MPT:
-
Methenyltetrahydromethanopterin
- M_N2 :
-
Nitrogen
- M_NADH:
-
NADH
- M_NADPH:
-
NADPH
- M_NH4 + :
-
Ammonium ions
- M_OAA:
-
Oxalacetate
- M_PEP:
-
Phosphoenolpyruvate
- M_Phe:
-
Phenylalanine
- M_PLIP:
-
Phospholipids
- M_PPA:
-
Prephenate
- M_Pro:
-
Proline
- M_PROT:
-
Protein
- M_PRPP:
-
Phosphoribosyl pyrophosphate
- M_PYR:
-
Pyruvate
- M_RIB5P:
-
Ribose-5-phosphate
- M_RNA:
-
RNA
- M_SED7P:
-
Sedoheptulose-7-phosphate
- M_Ser:
-
Serine
- M_SKA:
-
Shikimate
- M_SUCC_CoA:
-
Succinyl coenzyme A
- M_Thr:
-
Threonine
- M_Trp:
-
Tryptophane
- M_Tyr:
-
Tyrosine
- M_Val:
-
Valine
- M_X:
-
Biomass
- M_XYL5P:
-
Xylose-5-phosphate
Intracellular metabolites of C. butyricum
- P_13PD:
-
1,3-Propanediol
- P_3HPA:
-
3-Hydroxypropionealdehyde
- P_AcCoA:
-
Acetyl coenzyme A
- P_AKG:
-
α-ketoglutarate
- P_ATP:
-
ATP
- P_CO2 :
-
Dioxide
- P_DHA:
-
Dihydroxyacetone
- P_DHAP:
-
Dihydroxyacetonephosphate
- P_E4P:
-
Erythrose-4-phosphate
- P_EtOH:
-
Ethanol
- P_FdH:
-
Ferredoxin (reduced)
- P_FORM:
-
Formate
- P_FRU6P:
-
Fructose-6-phosphate
- P_GA3P:
-
Glyceraldehyde-3-phosphate
- P_GLC:
-
Glycerol
- P_GLU6P:
-
Glucose-6-phosphate
- P_H2 :
-
Hydrogen
- P_HAc:
-
Acetic acid
- P_HBu:
-
Butyric acid
- P_ISOCIT:
-
Isocitrate
- P_LAC:
-
Lactate
- P_NADH:
-
NADH
- P_NADPH:
-
NADPH
- P_NH4 + :
-
Ammonium ions
- P_OAA:
-
Oxalacetate
- P_PEP:
-
Phosphoenolpyruvate
- P_PYR:
-
Pyruvate
- P_RIBOS5P:
-
Ribose-5-phosphate
- P_RIBUL5P:
-
Ribulose-5-phosphate
- P_S7P:
-
Sedoheptulose-7-phosphate
- P_X:
-
Biomass
- P_XYL5P:
-
Xylose-5-phosphate
Rights and permissions
About this article
Cite this article
Bizukojc, M., Dietz, D., Sun, J. et al. Metabolic modelling of syntrophic-like growth of a 1,3-propanediol producer, Clostridium butyricum, and a methanogenic archeon, Methanosarcina mazei, under anaerobic conditions. Bioprocess Biosyst Eng 33, 507–523 (2010). https://doi.org/10.1007/s00449-009-0359-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-009-0359-0