Abstract
A kinetic metabolic model describing Catharanthus roseus hairy root growth and nutrition was developed. The metabolic network includes glycolysis, pentose-phosphate pathway, TCA cycle and the catabolic reactions leading to cell building blocks such as amino acids, organic acids, organic phosphates, lipids and structural hexoses. The central primary metabolic network was taken at pseudo-steady state and metabolic flux analysis technique allowed reducing from 31 metabolic fluxes to 20 independent pathways. Hairy root specific growth rate was described as a function of intracellular concentration in cell building blocks. Intracellular transport and accumulation kinetics for major nutrients were included. The model uses intracellular nutrients as well as energy shuttles to describe metabolic regulation. Model calibration was performed using experimental data obtained from batch and medium exchange liquid cultures of C. roseus hairy root using a minimal medium in Petri dish. The model is efficient in estimating the growth rate.
Similar content being viewed by others
Introduction
In vitro plant biotechnology offers a controlled environment and has been widely studied for phyto-pharmaceuticals and recombinant proteins production. However, the low productivity and the poor reproducibility of the cultures are still limiting the economical feasibility of such in vitro bioprocesses. Moreover, the lack of reproducibility of the cultures significantly complicates process validation and acceptance by the regulatory agencies, and thus the potential to rapidly put a product to market. Different approaches have succeeded in decreasing the technological risk associated with in vitro plant biotechnology. The introduction of elicitors such as chitin [1] and jasmonic acid [2] has shown to enhance significantly the production level for many plant species cultured as cell suspensions and hairy roots. The use of an extractive phase allowed the simplification of harvesting procedures [3, 4]. More recently, cell engineering studies have shown its potential in improving cell catalytic capacity towards the production of secondary metabolites [5–7] and in in vitro molecular farming for recombinant human proteins [8]. However, high variability levels in cell growth and production of biomolecules of interest are still observed. The genetic flexibility of plant cells may partly explain these phenomena [9] and inadequate culture management may also be involved.
A plant cell has the ability to accumulate nutrients and metabolites which are involved in the regulation of its metabolic pathways. The key for improving plant cell culture reproducibility may thus rely on a better understanding of the links that are exerting between a plant cell physiological state and its potential towards growth and production of a biomolecule of interest. This understanding (following that of Bailey [10]) could take the form of a descriptive and predictive metabolic model. Such structured model may then be either useful to enhance our understanding of cell behaviour, in identifying possible regulatory roles [11], as well as being a tool for defining adequate controlled culture conditions. Metabolic modelling has been applied to plant cells for studying specific metabolic sub-networks such as photosynthesis [12, 13], respiration [14], cellulose biosynthesis [15] and lipid biosynthesis [16]. These studies have clearly showed the importance of the energy shuttles on the control of the metabolic pathways. In addition, some nutrients are known to be involved in the regulation of the cell metabolism. Intracellular inorganic phosphate (Pi) plays a central role in the regulation of enzymes activity through phosphorylation/dephosphorylation processes, ATP/ADP concentration ratio, starch synthesis and storage, and in the flux distribution between the glycolysis and the pentose-phosphate pathway (PPP). Intracellular nitrate and ammonium are known to affect amino acid (AA) production.
The large capacity of plant cells for nutrient and metabolite accumulation plays a crucial role in cell growth and biomolecule production, as observed for Daucus carota hairy root [17] and Eschscholtzia californica suspension cells [18]. Recently, we have developed a kinetic model based on intracellular nutrients such as inorganic phosphate, nitrate and sugars which showed to be efficient in simulating carrot hairy root growth for different culture media composition [17]. Therefore, the aim of this work was to include the description of metabolic pathways to the nutritional model in order to describe plant cells behaviour from the estimation of the cells physiological state, including nutritional and metabolic states. Catharanthus roseus was studied as a model biological system. Cell nutritional state in Pi, nitrogen (NO −3 and NH +4 ) and carbohydrates (sucrose, fructose, glucose and starch) is described. The hypothesis of a central primary metabolism at steady state has been proposed based on literature [19, 20]. Using the metabolic flux analysis (MFA) approach, a model reduction [21] was applied on the central primary metabolism network and resulted in independent pathways. A second network includes transient fluxes such as for nutrient uptake and storage, energy shuttles management and root cells growth. Metabolic regulation of the fluxes from energy shuttles and nutrients is included. The hairy root specific growth rate is described as a function of the content in cell building blocks such as amino acids (including proteins), lipids (LIP), organic acids (ORA), organic phosphates (OP) (including nucleic acids) and structural hexoses (STH). Batch and medium exchange cultures of C. roseus hairy root were performed and the experimental data were used for model calibration.
Model general structure
The model has been first developed by Tikhomiroff [22]. The cell metabolic network (Fig. 1) is divided into two interlinked sub-networks as the stationary (SPMP) (Fig. 2) and the transient (TPMP) primary metabolic pathways (Fig. 1). The SPMP includes glycolysis, PPP, the TCA cycle and the catabolic reactions leading to the cell building blocks. The cell building blocks are amino acids and peptides which were taken as a unique pool of AA, ORA, OP, LIP and STH. The TPMP network is linked to the SPMP network and describes cells growth and nutrient transport between medium and intracellular volumes. Compartmentalization of nutrients and metabolites among the cytosol, the vacuole and other organelles is not included in the model and a single cell population was considered to describe the hairy root cells pools. This simplification already showed to be efficient to model hairy root growth and nutrition [17]. The secondary metabolism is simplified to fluxes leading to two pools, one accounting for the global pool in secondary metabolites derived from tryptamin (TRYSM), and one accounting for the global pool in secondary metabolites derived from secologanin (SECSM). The model is thus composed of interlinked metabolic networks that are at steady (SPMP) and transient (TPMP) states, and is described by a mass balance using the stoichiometric matrix and the hairy root specific growth rate:
where S is the stoichiometric matrix, Φ is a vector containing reaction fluxes, μ is the hairy root specific growth rate and M is a vector containing the concentration in cellular metabolites and nutrients. Root mass with time can then be estimated both kinetically and from a mass balance on all cell constituents.
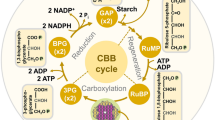
Fluxes in the stationary primary metabolic pathways (SPMP). Flux numbers refer to the stoichiometric biochemical reactions of Table 1
Stationary primary metabolic pathways
The pseudo-steady-state assumption for the central metabolism was based on observations from Rontein et al. [19] and Stitt and Fernie [20] and proposed to simplify the model development. The original SPMP metabolic network has 31 fluxes (Fig. 2; Table 1), which were reduced to 20 independent pathways (Fig. 3; Table 2) using the method proposed by Simpson et al. [21] and Stephanopoulos et al. [23] and the following simplifications. Briefly, the minimal number of independent feasible metabolic pathways is determined with a group of metabolites assumed at steady state: G6P, F6P, R5P, G3P, E4P, CHO, PEP, PYR, A-CoA, OXO and OAA. The “N” matrix (11×31) is first constructed from the 31 biochemical reactions and the 11 metabolites at steady state. The determination of the independent pathways then requires the knowledge of the kernel matrix “K” (31×number of independent pathways), which describe each pathway as a linear combination of the 31 biochemical reactions of the SPMP. This matrix is the non-trivial solution of the equation:
Since this equation has an infinite number of solutions, both “N” and “K” matrices are decomposed each into two matrices:
with “ N 1” and “ K 2” selected as square invertible matrices. Starting using identity matrix for “K 2”, “K 1” can then be determined by modifying “K 2” such that possible independent pathways can be identified:
Using the complete matrix K, we can build the independent pathways presented in Fig. 3. In this work, the size of this matrix is 31×20. The complete method to obtain the pathways can be found in Ref. [22].
Reduced independent biochemical pathways from the SPMP. Pathway numbers refer to the stoichiometric biochemical reactions of Table 2. Kinetic description of the resulting fluxes is presented in Table 4. The 20 independent biochemical pathways were obtained from the 31 biochemical reactions of the SPMP described in Fig. 2 and Table 1, using the reduction method of Simpson et al. [21] and Stephanopoulos et al. [23], as described in Sect. 3
The nutrient concentrations described are glucose (GLC), fructose (FRU), sucrose (SUC), ammonium (NH4) and inorganic phosphate (Pi). The anaplerotic pathways are simplified to the transformation of phosphoenolpyruvate (PEP) into oxaloacetate (OAA). The metabolite pools resulting from the SPMP are starch (STA), STH (cell wall and membrane constituents), OP (nucleotides, phospholipids, nucleic acids), total AA including that of peptides, ORA, pools from tryptamin (TRYSM) and from secologanin (SECSM). Starch biosynthesis (flux v(r14) in Table 2) and catabolism (v(r13)) are included. AA (v(r5)) are assumed to be synthesized from oxoglutarate, since it is the site for ammonium fixation and thus the initiation step in the AA biosynthesis [24], even though there are AA synthesized from other pathways. Tryptamin (TRYSM) biosynthesis (v(r1)) has been explicitly described because this AA is a precursor, with secologanin (SECSM) (v(r2) and v(r10)), to the formation of the secondary metabolites. OP biosynthesis (v(r11)) has been simplified as illustrated in Fig. 3 and is accounting for the four nucleotides. The biosynthesis of STH results from multiple biochemical reactions [25] but it is simplified to the condensation of G6P with R5P (v(r12)). The biosynthesis (v(r7)) and catabolism (v(r15)) of LIP is linked to acetyl-coenzyme A [26] as for ORA biosynthesis (v(r16)) and catabolism (v(r17)). The kinetic expressions of the fluxes are described in the next section.
Transient primary metabolic pathways
The TPMP network includes the biochemical reactions that cannot satisfy the pseudo-steady-state hypothesis. These include nutrients transport and accumulation, root cells growth and energy shuttles dynamics. The general structure of the TPMP and its interactions with the SPMP are illustrated in Figs. 1 and 2. The stoichiometric mass balances are presented in Table 3 and the kinetic terms used in fluxes’ regulation are described in Table 4. Extracellular nutrients such as sucrose (ESUC), glucose (EGLC), fructose (EFRU), ammonium (ENH4), nitrate (ENO3) and Pi (EPi) are included. Sucrose is hydrolysed into fructose and glucose extracellularly by apoplastic invertases (Table 3, v(28)), or intracellularly as described in the SPMP (Table 2, v(r19)). It should be noted that sucrose biosynthesis (Table 2, v(r20)) is not included into the model because there are no evidence that hairy roots have active chloroplasts. v(r19) then represents the net flux of sucrose hydrolysis. Alternative glycolytic pathways [27] are described and the carbohydrate sources include sucrose, glucose, fructose and starch. However, since all the biosynthesis pathways of the SPMP are defined with glucose as precursor, all available intracellular carbon sources have first to be converted into glucose. Fructose conversion (v(r3)), starch catabolism (v(r13)) and sucrose hydrolysis (v(r19)) processes are thus feeding the intracellular glucose pool. Plant cell adaptation mechanism to Pi deficiency is described with the fluxes for the degradation of pyrophosphate (v(26)) and OP (v(23)) into Pi. The plant cell storage capacity for sugars, NO −3 , NH +4 , Pi and cell building blocks is described. Respiration is described (v(30)), however, oxygen is assumed to be non-limiting as discussed below. Energy loss associated with maintenance and other reactions that are not included into the SPMP is contained in flux v(29).
Metabolic regulation
Metabolic regulation of two kinds is integrated into the model: that associated with the energetic status of the cells and that related to the nutritional state of the cells, variable denoted as “θ i ” below. Each flux (ν) is regulated as follows, according to a multiplicative kinetic of each mechanism involved.
The order of the regulation kinetics can also be adjusted from the term α i to account for multi-steps mechanisms. Since we established the independent pathways for the SPMP with arbitrary conditions, it is not possible to compare the structure of the resulting kinetics with literature. However, the identified independent pathways have been selected for their feasibility, and their combination is mathematically equivalent to the whole metabolic system (31 fluxes), given that the pseudo-steady-state hypothesis is acceptable.
Furthermore, a sigmoid function is used for nutrients acting like switches to avoid large discontinuities when solving the differential equations, which would have been the case using a simple on/off switch type. Continuous functions are also more representative of biological behaviour [28]. The sigmoid function is also used for imposing maximum accumulation levels (see Table 4):
The parameter “a” defines the steepness of the function, “M i ” is the concentration of the nutrient involved in the regulation of a flux and “M i,t ” is the nutrient concentration threshold.
The role of energy shuttles in the regulatory mechanisms is described in Table 4. NADH, NADPH and NTP (the sum of ATP, GTP, CTP and UTP) as well as their reduced forms (NAD, NADP and NDP) are used. The sum in energy shuttles per g DW of roots in both oxidized and reduced forms (NADH + NAD; NDP + NTP; NADPH + NADP) are taken constant with time. However, the energetic status of the cells [NADH/(NADH + NAD); NTP/(NDP + NTP); NADPH/(NADPH + NADP)] was not assumed constant. ATPase proton pumps that are linked to H+/nutrient co-transport and involved in the control of intracellular pH are thus included. ATP consumption for transmembrane transport of glucose (v(21)), fructose (v(22)), sucrose (v(24)), nitrate (v(27)) and Pi (v(31)) is thus included. A unique value for the affinity constant for NTP (K mNTP) is used. The same strategy of using unique affinity constants for NAD, NADH, NADP and NADPH is also applied.
Root cells nutritional state in Pi, NH4 and in sugars are involved in metabolic flux regulation. Intracellular Pi is involved in enzyme activation/deactivation processes through dephosphorylation/phosphorylation, in the biosynthesis of NTP (v(30)) and of many other metabolites as described below. Intracellular NH4 is involved in AA biosynthesis (v(r5)) (Fig. 3; Table 4). Michaelis–Menten kinetics with NH4 and Pi are at power two since a second-order mechanism is involved. NH4 has first to be transformed into NH2 radical, then into an AA which is finally integrated into a protein structure. Management of carbon source is crucial for plant cells since glucose, fructose, sucrose and starch can be used. However, since the model is based only on glucose, biochemical reactions converting other sugars into glucose are described (Fig. 3; Table 4). Starch biosynthesis (v(r14)) is controlled by the total intracellular available sugar concentration, and the sum in intracellular GLC, FRU and SUC has to be above 0.2 mmol/g DW for the STA biosynthesis to occur. A maximal STA storage capacity of 1.5 mmol/g DW was observed experimentally (see Sect. 7). Starch is degraded (v(r13)) when the GLC concentration is below 0.2 mmol/g DW. PPP (v(r4)) is regulated by NADP/(NADP+NAPH) ratio, which has to be above 0.5 for the pathway to be active. Pyruvate kinase regulation (v(r6)) is under the control of intracellular Pi concentration with a flux increase at 1 mmol Pi/g DW and below. G3P conversion into PEP (v(r8)) is regulated by Pi concentration with a flux increase at 1 mmol Pi/g DW and below. TCA cycle (v(r9)) is regulated by the NAD/(NAD+NADH) ratio, and a ratio that is above 0.5 will induce a flux increase. GLC (v(21)) and FRU (v(22)) uptake require a STA concentration that is below 0.07 and 0.02 mmol/g DW, respectively. OP synthesis (v(r11)) is regulated by intracellular Pi concentration with a flux increase at above 0.1 mmol Pi/g DW. OP degradation (v(23)) is controlled by intracellular Pi concentration with a flux increase at below 0.045 mmol Pi/g DW. LIP (v(r15)) and ORA (v(r17)) degradation into A-CoA is controlled by NADH concentration with fluxes increase at below 1 mmol NADH/g DW. Phosphofructokinase regulation (v(r18)) is controlled by intracellular Pi with a flux increase below 1 mmol Pi/g DW. Respiration rate (v(30)) is controlled by NDP/(NTP+NDP) ratio with a flux increase below a ratio of 0.5. Finally, regulation of root cells growth rate (v(32) is controlled by the intracellular concentration in the cell building blocks such as OP, LIP, amino acids and peptides (AA), ORA and STH. Monod model was used for LIP, AA and ORA. A hybrid Moser–Monod model was used for OP and STH with each kinetic term at power 4 and 1.25, respectively (Table 4). Since OP and STH are crucial to cell growth, the steepness of the affinity for both cell building blocks was increased.
Materials and methods
Culture conditions
Liquid cultures of hairy roots were performed in Petri dish as described in Jolicoeur et al. [17]. The major problem in culturing hairy roots is the difficulty to obtain a representative sample of the root network since the roots develop a highly dense interlinked bed. Petri dish culture allowed distributing the roots inoculum in a series of dishes with each dish taken as a single sample. Root growth was also limited by the use of a low salt medium, thus preventing the occurrence of dense root network. Previous results in Petri dish [17] and in bioreactor [3] suggest that there was no oxygen limitation in the cultures of this work.
Hairy roots of C. roseus L. G. Don were established as described by Bhadra et al. [29], with Agrobacterium rhizogenes strain A4. Hairy roots were transferred every month in Petri dishes in 25 mL of minimum medium [30] supplemented with 3% (w/v) sucrose and with a threefold KH2PO4 (0.352 mM). In the batch culture experiments, approximately 0.125 g fresh weight (FW) of hairy roots was inoculated in each Petri dish containing 25 mL of minimum medium. In the medium exchange cultures, approximately 0.125 g FW of hairy roots was inoculated in Petri dish containing 25 mL of minimum medium. The medium was renewed at 3- or 2-day intervals to avoid depletion of Pi in the medium. Whole Petri dishes were harvested in triplicates (n=3) periodically and taken as distinct samples. Liquid from each dish was filtered at 0.45 μm (Millipore, Billerica, Massachusetts) and stored at −20°C for further analysis. Roots were filtered under vacuum on a glass fiber filter (47 mm diameter Glass Microfiber filters GF/D, Whatman, #1823 047) and rinsed three times with 20 mL of de-ionized water. The filtered roots were removed from the filter and weighed for FW in a disposable aluminum dish (Fisher Scientific, # 08–732) on a precision balance (Sartorius). Fresh roots were immediately frozen into liquid N2 and stored in liquid N2 for further analysis. Root samples were then freeze dried (Duratop and Duradry, FTS Systems Inc., Stone Ridge, NY, USA), weighted for DW measurement and grinded (mortar and pestle) for further analysis. All further analyses were performed using freeze-dried roots.
Amino acids analysis
Approximately 2 mg of freeze-dried roots were extracted in 1 mL of 2% w/v 5-sulfosalicylic acid. The samples were sonicated for 15 min and then centrifuged at 12,000g for 5 min. The supernatant was analysed for AA by HPLC using a modified method from Gombert et al. [31] as described in Benslimane et al. [32]. The column is a high-efficiency Nova-Pak TM (C18, 4 μm). Precolumn derivatization of the AA was performed using AccQ.Fluor (6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, or AQC), which is an N-hydroxysuccinimide-activated heterocyclic carbamate, and the derivatized AA were quantified via a fluorescence detector. α-Aminobutyric acid was used as an internal standard. The standard error of measurement using this method was routinely below 5%. AA concentration was then calculated since the peak for each individual AA accounted for the combination of free and that from peptides and proteins.
Extraction of sugars
Approximately 10 mg of freeze-dried roots was washed with 80% ethanol and then centrifuged at 16,000g for 5 min. This wash was done three times and each time the supernatant was kept for glucose, fructose and sucrose analyses. After the third wash the pellet was kept for starch analysis.
Analysis of sugars
Glucose, fructose and sucrose analysis was performed with a Beckman Coulter HPLC (Beckman Coulter Canada Inc, Mississauga, Canada; Pump model 126, automatic injector model 508) equipped with a Gilson model 132 refractive index detector. A Prevail Carbohydrate ES column 4.6 mm×250 mm (Alltech Canada, Guelph, Ontario, Canada), coupled with a Prevail Carbohydrate ES All-Guard 4.6 mm×7.5 mm guard column (Alltech Canada, Guelph, Ontario, Canada), was used at a column temperature of 35°C. The injection volume was 20 μL. The mobile phase consisted of acetonitrile and water 75:25 (v/v) at 1.0 mL/min. The cell pellet obtained from the soluble sugars extraction was re-suspended in 1 mL de-ionized water and sterilized at 121°C for 15 min together with a 1 mL 6 g/L starch solution in de-ionized water. Samples were allowed to reach room temperature and their volume was readjusted to 1 mL with de-ionized water if necessary. Starch calibration standards from 0 to 6 g/L were prepared by serial dilutions of the starch solution in de-ionized water. Calibration standards and samples were diluted 1:1 with an amyloglucosidase solution (Sigma, St. Louis, Missouri, USA, Cat. #S9144) and incubated for 15 min in an ultrasound bath at 60°C. Samples and standards were centrifuged for 10 min at 16,000g. A 10 μL aliquot of the supernatant was directly transferred into a spectrometric cuvette with 500 μL of “Glucose Infinity” reagent (Sigma, Cat. #17-25). After incubation (15 min) at room temperature, 500 μL of 100 mM KH2PO4 buffer pH 7.5 was added followed by spectrophotometric reading at 340 nm.
Ions extraction
Ions were extracted from approximately 10 mg of freeze-dried roots in 1.5 mL of 5% (w/v) trichloroacetic acid. This mixture was sonicated at 40°C for 30 min and then centrifuged at 16,000g for 10 min. The supernatant was filtered at 0.45 μm and analysed by HPLC.
Ions analysis
Culture medium and intracellular contents in majors ions (Cl−, NO −3 , H2PO −4 , SO 2−4 , NH +4 , K+, Na+, Ca2+) were analysed using a Dionex HPLC system (Dionex Canada Ltd., Oakville, Canada) equipped with an isocratic pump, an automated sampler AS-3500 and a pulsed electrochemical detector in the conductivity mode, controlled by the Dionex A1-450 software for cations and the Dionex Peaknet software for anions. Anions were separated using a 4×250 mm IONPAC AS14A-SC analytical column, an IONPACAG14A-SC guard column and a ASRS-1 anion self-regeneration suppressor to improve the signal-to-noise ratio. The mobile phase consisted of an aqueous buffer of 2 mM Na2CO3/1 mM NaHCO3 solution flowing at a rate of 1.0 mL/min. Cations were separated using a 4×250 IONPAC CS-12 analytical column, a IONPAC CG-12 guard column and a CSRS-1 cation self-regenerating suppressor. The mobile phase was an aqueous 20 mM methanesulphonic acid solution flowing at a rate of 0.9 mL/min.
Model simulations
The model simulations were done using the Matlab software (The MathWorks Inc., Natick, MA, USA). The differential equations system was integrated through the Ordinary Differential Equation solver ode15s.m. The model error minimization was performed by means of manual and algorithm-based methods. The former was done to find good initial estimates based on literature (when available). The latter was then used to reduce the global error between the estimates and the experimental data. The algorithm used was the lsqcurvefit.m subroutine (Optimisation toolbox, Matlab) based on the Levenberg–Marquardt algorithm [33].
Results and discussion
Model calibration and determination of the kinetic parameters
Model calibration was performed using experimental data from C. roseus hairy root batch and medium exchange cultures. Each experiment (one batch and one medium exchange) was performed in triplicate. A total of 14 samples were taken during the batch experiment and 17 during the medium exchange. Parameter values were obtained from experimental data (maximum uptake rates, maximum growth rate, sucrose maximum hydrolysis rate and some of the maximal accumulation levels) and from literature, taking values from other plant species when unavailable for C. roseus (Tables 5, 6, 7). Adjustment of the unknown parameters as well the values taken from literature was performed manually in parallel using a non-linear least-square algorithm (lsqcurvefit) from Matlab software. The least-squares criterion was applied on all the data points (triplicates taken as mean in (M i )exp). The sum of squares were weighted as described below.
where\(e_{i} = \sum {\left[ {\frac{{(M_{i})_{{{\text{exp}}}} - (M_{i})_{{{\text{model}}}} }} {{\tfrac{1} {2}({\text{max}}(M_{i})_{{{\text{exp}}}} + {\text{min}}(M_{i})_{{{\text{exp}}}})}}} \right]}\)
Error terms were weighted using the maximum (max(M i )exp) and the minimum (min(M i )exp) experimental values (exp) from either batch or medium exchange cultures. This method was preferred to a method using experimental error because the experimental error was very high for some data points and varied with experimental data (Table 8). This method has also showed previously to perform adequately to calibrate a nutritional model on hairy root data [17]. All the experimental data points from batch and medium exchange culture were used for the non-linear least-square regression. The total number of experimental data was then of 411 with 13 different measurements for each of the 31 samples plus eight measurements for intracellular AA. Many set of parameters were tested as initial guesses to improve the fitting. A combination of parameter values (Tables 5, 6) showed to minimize the global error for combined data from batch and from medium exchange cultures, using a convergence criterion of 1×10−6 on the variation of the global error. From the calculation of R 2 values (Table 8) for each of the 28 state variables that were fitted (14 state variables in two experiments), it seems that the best fit is obtained for cells growth and nutrient transport and storage (NO3, Pi, NH4). However, the sensitivity analysis also suggests that some parameters can still be optimized (Fig. 4). A critical problem with such a descriptive model is a high number of parameters. The model has 35 maximum reaction rates and 26 affinity constants and the least-square regression was only performed on only 14 of the 31 state variables of the model. It is then possible that the identified solution corresponds to a local minimum. More measurements on metabolite concentration are thus necessary to improve the performance of the model and for being able to perform an adequate identifiability analysis on the parameters. In this context, the proposed model is probably over-parameterized. However, the goal of this work was not to establish a minimal model but a descriptive model that can be used as a tool useful at improving our understanding of plant cell culture.
Sensitivity analysis on model parameters for batch (solid bar) and medium exchange (empty bar) cultures. Relative change in error is calculated as described in Sect. 7.1. Parameter adjustment of +50% (a). Parameter adjustment of −50% (b). Parameters not shown have absolute relative error changes that are below 0.05
Simulation of the cells physiological state
Hairy root growth
The model allows estimating root growth from flux ν(32) as well as from the total estimated mass of the root cells constituents per dish (Σ M i on a mass basis), taking average molecular weights for the cell building blocks (Fig. 5). However, the total mass calculated at initial condition (t=0) was 25% lower than that of the inoculum with 0.0148 g DW per dish as compared to 0.0197 g DW per dish, respectively. The difference of −0.0049 g DW per dish can be explained from the molecular species that are present in the cells but which are not included in the model. Therefore, the estimation of root mass with time obtained from ν(32) was compared with that calculated from total cell mass, but adding a constant correction factor of +25%. Model simulations then describe hairy root growth for both batch and medium exchange cultures. Both root mass estimates from ν(32) and ΣM i are superimposed (Fig. 5). However, the sum of the molecular species is overestimating measured biomass at the end of batch culture because of cell accumulation in diverse compounds while growth slowed down. Surprisingly, both batch and medium exchange cultures behaved similarly with a maximum specific growth rate of 0.035 d−1 and a growth cessation at around 40 d. Periodic medium renewal has avoided nutrient depletion in macronutrients and sugars (Fig. 6). However, the plateau observed for the medium exchange culture may have resulted from a limitation in micronutrients, which were not measured. Therefore, only experimental data obtained before the occurrence of the plateau for the medium exchange culture were used for model calibration and other analyses. The strategy of describing cell growth as a function of intracellular content in building blocks thus showed to perform adequately. This result may support the global model structure as well as the hypothesis of pseudo-steady state for the SPMP. The central primary metabolic network, described here as SPMP, was observed at pseudo-steady state by Rontein et al. [19] for most of in vitro culture duration for tomato suspension cells [20]. However, this hypothesis has to be further investigated.
Model simulation for Catharanthus roseus hairy root batch (filled square) and medium exchange (triangle) liquid cultures in Petri dish. a–e. Intracellular concentration in cell building blocks with time. Batch (f) and medium exchange (g) cultures. The specific growth rate is described from ν(32) as a function of intracellular concentration in cell building blocks. Model simulations for batch (solid lines) and medium exchange (dashed lines) cultures, and total mass for the molecular species accounted in the model increased of 0.0049 g DW accounting for non-estimated molecular species (dashed line). Root mass was calculated using average MW for STA (180.15, based on glucose units); SEC (400); TRY (187); NAD/NADH (712/713); NADP/NADPH (744.4/745.4); NDP/NTP (476/507); ORA (809.75); STH (180.15, based on glucose units); OP (average of 305 from nucleotides); LIP (810, based on that of A-CoA); IPP (246) and AA (average of 136.75)
Extracellular concentration in nutrients with time. Experimental data for C. roseus hairy root batch (filled square) and medium exchange (triangle) liquid cultures. Concentration in sugars (a–c), inorganic phosphate (d) and nitrate (e) in the culture media with time. Model simulations for batch (solid lines) and medium exchange (dashed lines) cultures
The cells building blocks
The measured total AA concentration decreased with time for the batch culture and reached a plateau for the medium exchange culture (Fig. 5a). Simulated AA concentrations seem to follow the trend for experimental data but were overestimated for both batch and medium exchange cultures. The deviations cannot be explained, however, by exuded AA and proteins since no AA were detected in the used culture media. Values of 0.63±0.10 to 0.82±0.10 mmol AA/g DW were measured for the batch culture which corresponds to 0.088±0.014 and 0.12±0.014 g AA/g DW, respectively (taking an average amino acid MW of 140 g/mol). Higher values were measured in the medium exchange culture with 0.63±0.15 to 1.1±0.14 mmol AA/g DW, which represents 0.12±0.021 and 0.16±0.020 g AA/g DW, respectively. Free AA content of 0.21 mmol AA/g DW (estimated at 0.029 g AA/g DW) is reported in literature for tobacco suspension cells using B5 medium [34] and 0.15 mmol AA/g DW (estimated at 0.021 g AA/g DW) in potato tuber [35, 36]. In tomato cell culture, the total protein content was of 0.1–0.4 g proteins/g DW [19]. The total AA mass per root dry weight that have been measured, which represents the sum of free AA and that of peptides and proteins, is thus within the range reported in literature. The use of a minimal medium may explain a low value for total AA. The other cell building blocks were not measured but the model estimations were close to literature data. Total ORA concentration was estimated at 0.15 mmol/g DW as compared to 0.42 mmol/g DW reported by Farré et al. [35]. Total STH were estimated at 0.14 mmol/g DW as compared to a range of 0.3–1.6 mmol/g DW (on the basis of glucose units). Maximal simulated rates of biosynthesis were compared with that from literature. The rate of AA biosynthesis (v(r5)) was estimated at 0.12 mmol/g DW/d, which was within the range of 0.094–0.36 mmol/g DW/d reported by Rontein et al. [19] for tomato suspension cells. STH biosynthesis (v(r12)) was slow with 0.009 mmol/g DW/d as compared to 0.36 mmol/g DW/d [19]. Maximal net rate of starch (STA) biosynthesis (v(r14)) was 0.20 mmol/g DW/d as compared to 0.15 mmol/g DW/d [19]. Differences in metabolic fluxes may be explained from the differences in the specific growth rate. Rontein et al. [19] have reported a specific growth rate of 0.4 d−1 for tomato suspension cells in B5 medium as compared to 0.035 d−1 in this study for C. roseus using a minimal medium. The same minimal medium has been reported previously to induce lower growth rate as compared to standard media for carrot hairy root [17].
The cells nutritional state
Extracellular sucrose (ESUC) concentration profile was simulated by the model for both the batch and the medium exchange cultures (Fig. 6), thus suggesting that the sucrose hydrolysis modelling strategy was adequate. In the case of glucose (EGLC) and fructose (EFRU), simulations for the batch culture followed the trend of experimental data but before growth cessation. Then, estimated glucose and fructose levels stayed high as compared to experimental data. This may suggest that the model is underestimating glucose and fructose uptake at reduced growth. However, simulated concentrations in intracellular sucrose (SUC) and glucose (GLC) were following similar trends than experimental data (Fig. 7). However, model simulated constant starch (STA) concentration and overestimated fructose (FRU) concentration. In the case of the medium exchange culture the model simulated adequately experimental data on extracellular and intracellular sucrose and intracellular glucose. Extracellular glucose and fructose concentrations were underestimated and starch and fructose concentrations were overestimated. A maximal sucrose rate of hydrolysis (v(28)) of 0.72 mmol/g DW/d was estimated as compared to 1.7 mmol/g DW/d for D. carota hairy root using same minimal medium [17]. Contents of 0.25 mmol SUC/g DW (batch) and 0.7 mmol SUC/g DW (medium exchange) were obtained as compared to 0.05 mmol SUC/g DW [19], 0.26 mmol SUC/g DW [36] and 0.4 mmol SUC/g DW [34]. For free glucose, cell contents of 0.3–0.05 mmol GLC/g DW were obtained as compared to 0.6 mmol GLC/g DW [19], 0.24 mmol GLC/g DW [36] and 0.18 mmol GLC/g DW [34]. For fructose, cell contents of 0.1–0.25 mmol FRU/g DW were observed as compared to 0.009 mmol FRU/g DW [35], 0.21 mmol GLC/g DW [34] and 0.5 mmol GLC/g DW [19]. Starch accumulation reached 1.25 mmol STA (based on glucose)/g DW as compared to 0.055 [34] and 0.1–0.6 mmol STA (based on glucose)/g DW [19].
Intracellular concentration in nutrients and metabolites with time. Experimental data for C. roseus hairy root batch (filled square) and medium exchange (triangle) liquid cultures. Intracellular concentration in sugars (a–d), inorganic phosphate (e), nitrate (f), ammonium (g), and in secondary metabolites issued from tryptamin (h) and secologanin (i) with time. Model simulations for batch (solid lines) and medium exchange (dashed lines) cultures
Model simulations of extracellular Pi and NO −3 (ENO3) concentrations followed experimental data for the complete duration of the batch and the medium exchange cultures (Fig. 6). Hairy root growth (simulated and experimental) has ceased concurrently to ENO3 depletion suggesting this ion to be limiting in the batch culture. Intracellular Pi and NH4 were simulated (Fig. 7). It is interesting to note that the final intracellular Pi concentration in the batch culture seemed to reach a plateau at 0.05 mmol/g DW which was simulated. This plateau may be close to the Pi level that is essential to maintain the endogeneous metabolism [37]. A maximum Pi accumulation plateau was also observed for the medium exchange culture around 0.2 mmol/g DW. This value is close to the maximum value of 0.23 mmol/g DW measured for carrot hairy root [17, and references therein]. Maximum intracellular nitrate of 0.6 mmol NO3/g DW was measured for both cultures. Intracellular nitrate was kept constant for the medium exchange and decreased to its initial value for the batch. Intracellular ammonium reached a value of 0.12 mmol NH4/g DW for the medium exchange culture and decreased to 0.09 mmol NH4/g DW after 30 d. For the batch culture, intracellular ammonium reached a maximum value (0.08 mmol NH4/g DW) at day 5 and then decreased closely to its value at inoculation. The model overestimated intracellular NO3 concentration and simulated experimental intracellular concentrations in NH4 for both the batch and medium exchange cultures. A more precise description of the secondary metabolism and of the AA biosynthesis may be required to improve model simulation of nitrogenous compounds.
The cells energetic state
Model has simulated energy rich cells with high levels in NTP and NADH at exponential growth (Fig. 8). However, estimated cell content in NADPH showed an opposite trend with lower values at exponential growth. This result is interesting since NADPH is produced through the pentose-phosphate pathway, which is mostly active at secondary metabolism. The simulated cell energetic level decreased after 20 d at the initial value (inoculation) in batch culture and was constant for the medium exchange culture. Since the medium was renewed every 3 days for the medium exchange experiment, a pseudo-steady state was expected. NTP/(NTP+NDP) and NADH/(NAD+NADH) ratios of ∼75% have been reported for tobacco suspension cells [34]. Farré et al. [35] measured ratios of ∼73% for NTP/(NTP+NDP) in potato tubers. These authors have measured an NTP level of 0.00013 mmol/g DW as compared to 0.00143 and 0.00074 mmol/g DW that was obtained from simulations at exponential growth and stationary growth, respectively. We have recently measured by in vivo NMR ATP levels of 0.009 mmol/g DW at day 1 and 5, and of 0.0065 mmol/g DW at day 10 for suspension cells of E. californica cultured using B5 medium [38]. Energetic levels simulated by the model were then similar to that found in literature. However, variation of energy shuttles concentration has to be further investigated.
Conclusion
A metabolic model capable of describing hairy root growth from the estimation of the cells physiological state was developed. The model includes the central metabolism, the primary metabolic pathways (SPMP) assumed at steady state and a network for the TPMP. At this point, the model is simplified and intracellular compartmentalization processes into the different cell compartments and organelles are not included. Nevertheless, the metabolic model showed to perform efficiently in simulating hairy root growth and nutrition. The use of intracellular concentrations in nutrients and co-substrates as well as the cells energetic state seems an efficient strategy in describing regulation of the metabolic fluxes. However, more experimental is required for improving the model structure as well as parameter values which may be regulated with cell physiological state. Finally, the model will be applied to other plant species as well as cell suspensions, and it will be studied as a tool to describe transient processes such as metabolic regulation.
Abbreviations
- a :
-
steepness of the function in Eq. 2
- AA:
-
amino acids (mmol/g DW)
- CHO:
-
chorismate (mmol/g DW)
- DW:
-
dry weight (g)
- e i :
-
normalized error of the substrate “i”
- EPi:
-
extracellular inorganic phosphate (mM)
- FW:
-
fresh weight (g)
- M i(t)exp :
-
nutrient or metabolite “i” concentration at time t from experimental data (mM or mmol/g DW)
- M i(t)model :
-
nutrient or metabolite “i” concentration at time t obtained by simulation (mM or mmol/g DW)
- \(K_{{{\text{m}},i}}\) :
-
affinity constant of the substrate “i” in Eq. 2 (mM or mmol/g DW)
- LIP:
-
lipids (mmol/g DW)
- M i :
-
nutrient or metabolite “i” concentration (mM or mmol/g DW)
- M i,t :
-
threshold concentration of a substrate in Eq. 2 (mmol/g DW)
- MFA:
-
metabolic flux analysis
- NDP:
-
nucleoside diphosphate (mmol/g DW)
- NTP:
-
nucleoside triphosphate (mmol/g DW)
- OP:
-
organic phosphate (mmol/g DW)
- ORA:
-
organic acids (mmol/g DW)
- Pi:
-
inorganic phosphate (mmol/g DW)
- PPi:
-
pyrophosphate (mmol/g DW)
- S i :
-
stoichiometric coefficient
- SEC:
-
secologanin (mmol/g DW)
- SPMP:
-
stationary primary metabolic pathways
- STA:
-
starch (mmol/g DW)
- STH:
-
structural hexoses (mmol/g DW)
- TPMP:
-
transient primary metabolic pathways
- TRY:
-
tryptamin (mmol/g DW)
- TSMP:
-
transient secondary metabolic pathways
- V :
-
liquid medium volume (L)
- X :
-
biomass (g DW)
- αi :
-
order of the regulation kinetic
- Φ i :
-
reaction flux i (mmol/d or mmol/g DW/d)
- ν(x):
-
reaction rate for the flux x (mmol/d or mmol/g DW/d)
- νGLC :
-
partial reaction rate for glucose (mmol/g DW/d)
- ν max(x):
-
maximum reaction rate for flux x (mmol/g DW/d)
- ν maxGLC :
-
maximum partial reaction rate for glucose (mmol/g DW/d)
- μ:
-
root cells specific growth rate (1/d)
- θ i :
-
nutrient, metabolite or energy shuttle “i” concentration (mmol/g DW)
References
Williams RD, Chauret N, Bédard C, Archambault J (1992) Effect of polymeric adsorbents on the production of sanguinarine by papaver somniferum cell cultures. Biotechnol Bioeng 40:971–977
Rijhwani SK, Shanks JV (1998) Effect of elicitor dosage and exposure time on biosynthesis of indole alkaloids by Catharanthus roseus hairy root cultures. Biotechnol Prog 14:442–449
Tikhomiroff C, Allais S, Klvana M, Hisiger S, Jolicoeur M (2002) Continuous selective extraction of secondary metabolites from Catharanthus roseus hairy root cultures with silicon oil in a two-liquid phase bioreactor. Biotechnol Prog 18:1003–1009
Byun SY, Pedersen H (1994) Two-phase airlift fermentor operation with elicitation for enhanced production of benzophenanthridine alkaloids in cell suspensions of E. californica. Biotechnol Bioeng 44:14–20
Lessard PA, Kulaveerasingam H, York GM, Strong A, Sinskey AJ (2002) Manipulating gene expression for the metabolic engineering of plants. Metab Eng 4:67–79
Verpoorte R, Contin A, Memelink J (2002) Biotechnology for the production of plant secondary metabolites. Phytochem Rev 1:13–25
Hughes EH, Shanks JV (2002) Metabolic engineering of plants for alkaloid production. Metab Eng 4:41–48
Sharp JM, Doran PM (2001) Strategies for enhancing monoclonal antibody accumulation in plant cell and organ cultures. Biotechnol Prog 17:979–992
Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18:1157–1161
Bailey JE (1998) Mathematical modeling and analysis in biochemical engineering: past accomplishments and future opportunities. Biotechnol Prog 14:8–20
Wiechert W (2002) Modeling and simulation: tools for metabolic engineering. J Biotechnol 94:37–63
Farquhar GD, von Caemmerer S, Berry JA (2001) Models of photosynthesis. Plant Physiol 125:42–45
Fridlyand LE, Scheibe R (1999) Regulation of the Calvin cycle for CO2 fixation as an example for general control mechanisms in metabolic cycles. BioSystems 51:79–93
Affourtit C, Krab K, Moore AL (2001) Control of plant mitochondrial respiration. Biochim Biophys Acta 1504:58–69
Delmer DP, Haigler CH (2002) The regulation of metabolic flux to cellulose, a major sink for carbon in plants. Metab Eng 4:22–28
Ramli US, Baker DS, Quant PA, Harwood JL (2002) Regulation of fatty acid synthesis. Biochem Soc Trans 30:1043–1046
Jolicoeur M, Bouchard-Marchand E, Bécard G, Perrier M (2002) Regulation of mycorrhizal symbiosis: development of a structured nutritional dual model. Ecol Model 158:121–142
Lamboursain L, Jolicoeur M (2002) In Vitro production of secondary metabolites by plant cells: the crucial role of the cell nutritional. In: Vasil IK (ed) Proceedings of the 10th international association for plant tissue culture and biotechnology congress. Kluwer, Dordrecht, pp 491–495
Rontein D, Dieuaide-Noubhani M, Dufourc EJ, Raymond P, Rolin D (2002) The metabolic architecture of plant cells: stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 277:43948–43960
Stitt M, Fernie AR (2003) From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon–nitrogen interactions. Curr Opin Biotechnol 14:136–144
Simpson TW, Follstad BD, Stephanopoulos G (1999) Analysis of the pathway structure of metabolic networks. J Biotechnol 71:207–223
Tikhomiroff C (2001) Étude des métabolismes primaire et secondaire des racines transformées de Catharanthus roseus en vue du développement d’une modèle cinétique. Master’s thesis. École Polytechnique de Montréal. 268 pp
Stephanopoulos GS, Aristidou AA, Nielsen J (1998) Metabolic engineering. Principles and methodologies. Academic, London, 725 pp
Michal G (1999) Biochemical pathways. Wiley, New York, 277 pp
Reid JS (1997) Carbohydrate metabolism: structural carbohydrates. In: Dey PM, Harbone JB (eds) Plant biochemistry. Academic, London, pp 205–236
Harwood JL (1997) Plant lipid metabolism. In: Dey PM, Harbone JB (eds) Plant biochemistry. Academic, London, pp 237–272
Plaxton WC (1998) Metabolic aspects of phosphate starvation in plants. In: Deikamn J, Lynch J (eds) Phosphorus in plant biology: regulatory roles in molecular, cellular, organismic, and ecosystem processes. American Society of Plant Physiologists, Rockville, pp 164–176
Thornley JHM, Johnson IR (1990) Plant crop modeling—a mathematical approach to plant and crop physiology. Oxford University Press, New York, 669 pp
Bhadra R, Vani S, Shanks JV (1993) Production of indole alkaloids by selected hairy root lines of Catharanthus roseus. Biotechnol Bioeng 41:581–592
Bécard G, Fortin JA (1998) Early events of vesicular–arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108:211–218
Gombert AK, Moreira Dos Santos M, Christensen B, Nielsen J (2001) Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J Bacteriol 183:1441–1451
Benslimane C, Elias CB, Hawari J, Kamen A (2004) Insights into the central metabolism of Spodoptera frugiperda (Sf-9) and Trichoplusia ni BTI-Tn-5B1–4 (Tn-5) insect cells by radiolabeling studies. Biotechnol Prog 21:78–86
More JJ (1977) The Levenberg–Marquardt algorithm : implementation and theory. Numerical analysis. In: Watson GA (ed) Lectures notes in mathematics 630:105–116
Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001) Fructose 2,6-biphosphate activates pyrophosphate: fructose6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212:250–263
Farré EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127:685–700
Roessner U, Wagner C, Kopka J, Tretheway RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. Plant J 23:131–142
Mimura T (1999) Regulation of phosphate transport and homeostasis in plant cells. Int Rev Cytol 191:149–200
Gmati D, Chen J, Jolicoeur M (2004) Development of a small-scale bioreactor: application to in vivo NMR measurement. Biotechnol Bioeng 89:138–147
Taiz L, Zeiger E (1998) Plant physiology. Sinauer Associates Inc., Sunderland, 690 pp
Brownleader MD, Harbone JB, Dey PM (1997) Carbohydrate metabolism: primary metabolism of monosaccharides. In: Dey PM, Harbone JB (eds) Plant biochemistry. Academic, London, pp 111–141
Contin A, van der Heijden R, Lefeber AWM, Verpoorte R (1998) The iridoid glucoside secologanin is derived from the novel trios phosphate/pyruvate pathway in a Catharanthus roseus cell culture. FEBS Lett 434:413–416
Tanner W, Caspari T (1996) Membrane transport carriers. Annu Rev Plant Physiol Plant Mol Biol 47:595–626
Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3:389–395
Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50:665–693
Cerezo M, Garcia AP, Serna MD, Primo-Millo E (1997) Kinetics of nitrate uptake by citrus seedlings and inhibitory effects of salinity. Plant Sci 126:105–112
Filleur S, Daniel-Vedele F (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207:461–469
Sakano K, Yazaka Y, Okihara K, Mimura T, Kiyota S (1995) Lack of control in inorganic phosphate uptake by Catharanthus roseus (L.) G. Don cells. Plant Phyiol 108:295–302
Schmidt ME, Heim S, Wylegalla C, Helmbrecht C, Wagner K (1992) Characterization of phosphate uptake by suspension cultured Catharanthus roseus cells. J Plant Physiol 140:179–184
Furihata T, Suzuki M, Sakurai H (1992) Kinetic characterization of two phosphate uptake systems with the different affinities in suspension-cultured Catharanthus roseus protoplasts. Plant Cell Physiol 33:1151–1157
Shimogawara L, Usuda H (1995) Uptake of inorganic phosphate by suspension-cultured tobacco cells: kinetics and regulation by Pi starvation. Plant Cell Physiol 36:341–351
Ullrich-Eberius CI, Novacky A, van Bel AJE (1984) Phosphate uptake in Lemna gibba G1: energetics and kinetics. Planta 161:46–52
Sentenac H, Grinon C (1985) Effect of pH on orthophosphate uptake by corn roots. Plant Physiol 77:136–141
Smith FA (1966) Active phosphate uptake by Nitella translucens. Biochim Biophys Acta 126:94–99
Beever RE, Burns DJW (1980) Phosphorus uptake, storage and utilization by fungi. Adv Bot Res 8:127–219
Mettler IJ, Leonard RT (1979) Ion transport in isolated protoplasts from tobacco suspension cells. II. Selectivity and kinetics. Plant Physiol 63:191–194
Bieleski R (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol 24:225–252
Acknowledgements
C. Tikhomiroff has a fellowship from the Fondation de l’École Polytechnique (France). Authors are grateful to A. Fazzi and S. Hisiger for HPLC analysis and to L. Bisson from BRI for the analysis of amino acids by GC–MS. The Canada Research Chair Program, NSERC and FQRNT funded this research project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Leduc, M., Tikhomiroff, C., Cloutier, M. et al. Development of a kinetic metabolic model: application to Catharanthus roseus hairy root. Bioprocess Biosyst Eng 28, 295–313 (2006). https://doi.org/10.1007/s00449-005-0034-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-005-0034-z