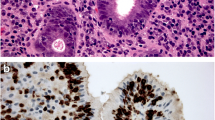
Abstract
The regenerating islet-derived (REG) gene family encodes a group of proteins highly expressed in several human pathologies, many of which are associated with epithelial inflammation. All human family members, namely REG1A, REG1B, REG3A and REG4, are closely related in genomic sequence and all are part of the c-type lectin superfamily. REGs are highly expressed during inflammatory bowel disease (IBD)-related colonic inflammation and the in vivo expression pattern of REG1A and REG4 has been localised by using immunohistochemistry. However, the function of the encoded proteins is largely unknown and the cellular localisation of REG expression during colonic inflammation has not been described. Therefore, we have used in situ hybridisation to demonstrate the cellular localisation of REG expression in healthy and diseased colonic mucosa. Samples drawn from an IBD cohort including both inflamed and un-inflamed colonic mucosa are described, as are samples from non-IBD inflammation and healthy controls. Immunohistochemistry against known cell-type markers on serial sections has localised the expression of REGs to metaplastic Paneth cells (REG1A, REG1B and REG3A) and enteroendocrine cells (REG4), with a marked expansion of expression during inflammation. The group of REGs can, based on gene expression patterns, be divided into at least two groups; REG1A, REG1B and REG3A with their expression focused in the crypt base spreading from Paneth cells and REG4 being more highly expressed towards the luminal face. This exploration of expression pattern forms provides the background for further exploration of REG function in the intestine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the discovery of the regenerating islet-derived (REG) gene family in connection with islet regeneration after experimental injury to rat pancreas, several studies have explored the possible functions and expression patterns of the various genes in the family. In the beginning, these studies focused mainly on the role of REG in ailments affecting the pancreas, such as diabetes, acute pancreatitis and pancreatic cancer. Today, we know that REG family proteins are highly expressed in the epithelial lining of many organs in connection with tissue injury and inflammation.
In humans, the REG gene family currently consists in four well-established members: REG1A, REG1B, REG3A and REG4. All are found encoded in tandem on chromosome 2p12, except for REG4, which is found on 1p12. The intron-exon arrangement of all REGs are similar, again except for REG4, which has considerably longer intronic sequences (Zelensky and Gready 2005). The protein sequence suggests similarities to the C-dependent lectins, although the carbohydrate-binding motif of the REGs does not display the calcium-binding site common for other members of the superfamily. Our current knowledge of human REG protein function and its expression pattern have recently been reviewed by Parikh et al. (2012).
The suggested functions of the various REGs are many and surprisingly diverse, given the great similarity in the sequence of the members. Of the human REGs, REG1A, REG3A and REG4 are the best studied. All REGs are expressed in the pancreas and small intestine, whereas only REG4 is constitutively expressed in colon epithelia (Zhang et al. 2003). During epithelial disease, in either pancreatitis or inflammation of the bowel, the expression of REGs increases. Both high REG4 and REG1A expression has been linked to poor prognosis in colorectal cancer (Astrosini et al. 2008; Oue et al. 2007) and both have been shown to have either a trophic or an anti-apoptotic effect. The effect of REG4 has been further investigated by using cancer cell lines and the results suggest that REG4 can stimulate both cell migration and proliferation (Guo et al. 2010; Rafa et al. 2010). Sekikawa et al. (2010) have demonstrated that the increase in REG1A expression seen in inflamed colonic mucosa correlates with IL22 expression and is stimulated in a STAT3-dependent manner. An antimicrobial effect has been demonstrated for murine Reg3g and human REG3A and is possibly specific for membrane-bound peptidoglycans of Gram-positive bacteria (Cash et al. 2006).
REG1A, REG1B, REG3A and REG4 all show increased expression during inflammatory bowel disease (IBD)-related inflammation of the colon (Dieckgraefe et al. 2002; Granlund et al. 2011; Lawrance et al. 2001). IBD is a group of chronic diseases affecting as many as 2.2 million Europeans and 1.4 million US citizens (Loftus 2004). The two main IBD subtypes, ulcerative colitis (UC) and Crohn’s disease (CD), are clinically distinct. They do, however, share many features such as chronic mucosal inflammation and epithelial injury. Despite the initial discovery of the high expression of REGs during IBD inflammation, few studies have investigated their expression pattern in the inflamed bowel mucosa. Dieckgraefe et al. (2002) have described REG1A in IBD by using immunohistochemistry (IHC); the protein is expressed by metaplastic Paneth cells of the inflamed colonic mucosa. In a comprehensive study by Oue et al. (2005), the expression patterns of REGIV (REG4) in many neoplastic and non-neoplastic tissues have been determined, with REG4 protein expression in normal colonic tissue being shown to be limited to enteroendocrine cells.
In a previous study, we have found REG mRNAs to be strongly up-regulated during inflammation of the colon. We have further examined, using IHC, the expression pattern of REG1A and REG4 in both in the un-inflamed and inflamed colonic mucosa from UC and CD patients, in normal colonic mucosa and in inflamed mucosa from patients with non-IBD antibiotic-induced pseudomembranous colitis (PC; Granlund et al. 2011). Whereas no REG1A expression is seen in normal mucosa, the Paneth cells found both in un-inflamed and inflamed IBD tissue and in inflamed tissue from patients with PC express REG1A protein. REG4 has been seen in serotonin-positive cells in all colonic samples. During inflammation, the expression of both REG1A and REG4 shifts to include all other epithelial cells of the colonic crypts, with expression for REG4 increasing towards the lumen and for REG1A increasing towards the crypt base.
Present knowledge of the mRNA expression patterns of the various REGs in human colonic mucosa is extremely limited, with only a few studies demonstrating the expression of REG1A and REG4 in colorectal cancer and pseudomyxoma peritonei of the appendix (Astrosini et al. 2008; Heiskala et al. 2006). In the present study, we have utilised an in situ hybridisation (ISH) method involving branched-chain DNA technology, enabling us, for the first time, to localise all human REGs in normal and inflamed colon. Protein expression has been studied with regard to the REGs for which antibodies are available. The expression pattern has been further investigated by identifying cell types expressing the various REGs by using IHC against appropriate cell-type markers in serial sections. The current study represents a comprehensive evaluation of REG expression pattern in the healthy and inflamed colon and will provide the necessary background information in further research on the regulation and function of these intriguing peptides.
Materials and methods
Samples
Sample material was drawn from a research biobank of IBD samples at St. Olav’s University Hospital, except for samples of PC, which were taken from the clinical files at the Department of Pathology, St. Olav’s University Hospital. Samples for IHC, ISH and microarray analysis were prepared as described previously (Granlund et al. 2011). In total, six groups of samples were defined: normal mucosa from healthy individuals (N), un-inflamed mucosa from the hepatic flexure (HF) in UC patients (UCU), un-inflamed mucosa from HF in CD patients (CDU), inflamed colonic mucosa from UC patients (UC), inflamed colonic mucosa from CD patients (CD) and inflamed mucosa from PC patients (PC). For the IBD samples and normal controls, the procedure was as follows (in brief). Four samples from the same area were collected from each patient by pinch biopsy during colonoscopy. Three were snap-frozen and further stored in liquid nitrogen, with one biopsy being fixed in formalin and subsequently embedded in paraffin. All un-inflamed biopsies from IBD patients and normal samples from healthy controls were taken from the hepatic flexure. Diseased samples were taken from areas in which the mucosa was macroscopically maximally inflamed. Hematoxylin-eosin (H-E)-stained sections from all samples were evaluated by an expert pathologist for verification of the diagnosis. Samples for which microscopic evaluation diverged from the initial endoscopic diagnosis were removed from the sample pool. Informed written consent was obtained from all study participants. The study was approved by The Regional Medical Research Ethics Committee (no. 5.2007.910) and was registered in the Clinical Trials Protocol Registration System (identifier NCT00516776).
Microarray gene expression analysis
Samples were prepared and data were analysed as described previously (Granlund et al. 2011). In brief, all RNA samples were controlled for RNA integrity and purity. Samples not fulfilling the quality requirements were removed prior to microarray analysis. In addition to the technology-specific quality controls supplied by Illumina, two samples were used as technical controls. One of these samples was hybridised to each 12-plex chip in order to detect any deviation in results. All data from the microarray analysis were analysed by using Bioconductor in the R statistical computing environment (Gentleman et al. 2004). In short, data were log-transformed and quantile-normalised and differential expression was identified by using limma linear regression (Smyth 2005). All P-values were corrected for multiple comparisons by using the Benjamini-Hochberg correction. The significance level was set to a corrected P-value of <0.05. All data from this analysis are available in ArrayExpress E-MTAB-184. A verification of the observed regulation of REG family genes by quantitative reverse transcription plus the polymerase chain reaction is given in a previous publication by Granlund et al. (2011).
Histological and immunohistological examination
Paraffin-embedded samples were cut into sections of 4 μm in thickness and mounted on glass slides. Three PC samples and five samples from each IBD sample group and normal controls were randomly selected. One section from each sample was stained with H-E for the evaluation of inflammatory status by an experienced pathologist. From each group, two samples displaying good crypt morphology were selected based on the evaluation of H-E-stained sections. These samples then formed the basis for the further evaluation of REG expression. This process was repeated twice, giving a total number of evaluated colonic samples of 12. The images shown in Results are all based on the six samples given in Table 1. For IHC and ISH, sequential pairs of sections were mounted with adjoining sides facing up in order to optimise serial section staining. Antibodies for REG1A (cat. no. RD181078100, BioVendor, Midrice, Czech Republic; dilution 1:500), REG4 (REGIV, cat. no. AF1379, R&D Systems, Minneapolis, Minn., USA; dilution 1:400), REG3A (cat. no. AB134209, Abcam, Cambridge, UK; dilution 1:100), Defensin alpha 6 (DEFA6, cat. no. HPA019462, Sigma, St. Louis, Mo., USA; dilution 1:1000) and serotonin (5HT, cat. no. AB16007, Abcam; dilution 1:200) were all commercially available. REG1A, REG3A, DEFA6 and serotonin were detected by using the Dako EnVision peroxidase kit (Dako, Glostrup, Denmark), whereas REGIV (REG4) was detected by using the Vectastain ABC kit (Vector Laboratories, Burlingame, Calif., USA). Dako DAB+ (diaminobenzidine) was used as the chromogene for all IHC, with all sections being counterstained with hematoxylin. Samples were de-paraffinised in Neo-Clear (Merck Millipore, Darmstadt, Germany) and antigen retrieval was performed by boiling sections in either sodium citrate (pH 6.0) or TRIS-EDTA (pH 9.0) buffer for 15 min. For ISH of REG1A, REG1B, REG3A, REG4 and Chromogranin A (CgA), the RNAscope 2.0 single-plex hybridisation system (Advanced Cell Diagnostics, Hayward, Calif., USA) was used according to the manufacturer’s recommendations. This method utilises branched-chain DNA technology to ensure good specificity and sensitivity (Player et al. 2001). In brief, tissue sections were heated (1 h, 60 °C) prior to de-paraffinisation and rehydration, followed by a protease treatment (30 min, 42 °C in a humidifying chamber). Each slide was hybridised with target probe for 1 h at 40 °C, followed by a series of specific signal amplification steps as described in the manufacturer’s protocol and was stained for 10 min by using DAB provided by the manufacturer. All sections were counterstained with hematoxylin. Initial hybridisation for each probe was performed including positive and negative controls in order to ensure proper target probe specificity. Control probes were provided by Advanced Cell Diagnostics. Positive controls targeted ubiquitin C, whereas negative control probes targeted bacterial dapB. All samples used in the final analysis performed as expected when hybridised with positive and negative control probes. Examples of positive and negative controls are given in Fig. S1.
All images were acquired by using NIS-Elements BR ver. 3.00 (Nikon Instruments, Melville, N.Y., USA). The white balance was automatically adjusted for each picture by means of the routines provided. Images were saved as TIFF-files. Subsequent image orientation and cropping was used to ease interpretation of serial sections.
Results
Samples
After evaluation of morphology, two samples from each group were selected for initial ISH. Fixation time for the samples varied and was unknown. As fixation time influences ISH strongly, this initial analysis was performed to ensure compatibility of the sample with the ISH protocol. The samples showing consistent results were used in the subsequent analysis. Sample information is given in Table 1. All samples used in the final analysis were tested with positive and negative control probes supplied by the manufacturer of the ISH kit. Quality controls showed minimal un-specific binding of the negative control probe and good hybridisation of the positive control probe. The analysis was repeated twice with separate sets of samples in order to confirm findings.
Microarray analysis
Results from the full microarray gene expression analysis have been described in previous publications (Granlund et al. 2011). All REG mRNAs were potently regulated in inflamed mucosa from UC and CD and were among the top eight differentially expressed genes for the contrast UC vs. N. The analysis did not show significant regulation of REG genes in un-inflamed samples but a subset of samples showed an expression level similar to that seen in inflamed samples.
For the present study, the REG expression levels for the randomly selected samples used in IHC/ISH analysis were extracted from the full set of microarray results. The LOG2 expression levels of the present samples are given in Table 2. The normal control had REG levels close to background, except for REG4, which displayed a basal expression slightly over background. The CDU sample exhibited a slight increase in the expression of all REGs, whereas the UCU sample had expression levels similar to inflamed samples thereby reflecting the dichotomous distribution of REG expression in un-inflamed IBD samples.
ISH and IHC analyses
Normal samples showed no ISH staining for REG1A, REG1B and REG3. For REG4, scattered cells showed positivity and were identified as serotonin-positive enteroendocrine cells. All IBD samples and PC samples showed signs of Paneth cell metaplasia as defined by DEFA6-positive cells in the crypt base. Figure 1 displays a selection of serial sections stained by IHC for DEFA6 and serotonin, with parallel ISH for REG1A, REG1B, REG3A and REG4.
Serial sections of colonic biopsy samples for in situ hybridisation (ISH) of REGs (a–e) with corresponding immunohistochemistry (IHC) for cell-type markers DEFA6 (e–g) and serotonin (h). Metaplastic Paneth cell (arrows in a-c, e-g). Enteroendocrine cell (arrows in d, h) Bar 30 μm. Sections a-c and e-g from un-inflamed ulcerative colitis. Sections d and h from pseudomembraneous colitis
The results from ISH of REG1A, REG1B, REG3A and REG4 in IBD samples are given in Fig. 2. During inflammation, the expression of all REGs increased. REG1A, REG1B and REG3A expression expanded upwards from the crypt base, whereas REG4 expression was higher towards the luminal end of the crypts, often with no expression in the lower third. During inflammation, all REGs appeared to be expressed by both absorptive colonocytes and goblet cells. The expansion of REG1B and to some degree of REG3A mRNA expression during inflammation, seemed limited compared with that for REG1A, rarely stretching much beyond the bottom third of the crypts. REG1A mRNA, on the other hand, could in some instances be found up to almost the luminal face, although the expression was limited to the lower half in most crypts. In un-inflamed samples, epithelial REG1A expression seemed to be limited to selected crypts. By closely inspecting serial sections, one could observe a correlation between the presence of Paneth cells and epithelial REG expression; a typical example is shown in Fig. 3. In some crypts of un-inflamed IBD mucosa and in all inflamed samples, REG4 expression was apparent in epithelial cells. The expression was strongest towards the luminal face of the sections. REG4 positivity was also seen in scattered enteroendocrine cells in samples with no REG4 positivity in epithelial cells. Figure 4 presents serial sections displaying these cells, demonstrating serotonin positive enteroendocrine cells expressing REG4.
ISH of all human REGs in colonic inflammatory bowel disease tissue. Sections are taken from samples representing un-inflamed ulcerative colitis (a–d), un-inflamed Crohn’s disease (e–h), inflamed ulcerative colitis (i–l) and inflamed Crohn’s disease (m–p). ISH was performed with probes for REG1A, REG1B, REG3A and REG4. Sample details are given Table 1. Bar 100 μm
Serial sections of a sample of un-inflamed mucosa from the hepatic flexure of a patient with Crohn’s disease. IHC for serotonin (a) and ISH for REG4 (b) and CgA (c) expression (CGA chromogranin A). A clear overlap of expression can be seen for all three markers, with the number of positive cells being highest for CgA and the lowest for REG4
A comparison of REG1A, REG3A and REG4 gene expression (as shown by ISH) and REG protein expression (as shown by IHC) in PC is given in Fig. 5. We found a good correlation between the extent of gene expression and protein expression. The sections used in Fig. 5 are subsequent sections, not serial sections as used in Fig. 1, resulting in poorer overlap between sections. The gene expression pattern for REG1A, REG3A and REG4 was similar to that seen in the IBD samples (results not shown).
Discussion
The colon mucosa is a rapidly renewing cell population with most cells undergoing a pattern of crypt-base proliferation, migration towards the lumen during differentiation and then apoptosis and shedding on the luminal surface within 1–2 weeks. In IBD colonic mucosa, crypt elongation is often observed, with an increase in both proliferation and apoptosis (Arai et al. 1999). In addition to the general morphological change with leucocyte infiltration being found in IBD, a change occurs in the cellular composition of the IBD colonic mucosa with, for example, Paneth cell metaplasia being seen in both the inflamed and un-inflamed IBD colon (Haapamäki et al. 1997; Lala et al. 2003). Changes also take place in the number and function of serotonin-positive enteroendocrine cells in the inflamed mucosa (Lomax et al. 2006). Our analysis shows that metaplastic Paneth cells in both un-inflamed samples from IBD patients and in inflamed colonic mucosa express REG1A, REG1B and REG3A mRNA. The main role of Paneth cells in their normal localisation, the mucosa of the small bowel, is to maintain homeostasis by controlling small intestinal microbial load through the secretion of antimicrobial peptides (Bevins and Salzman 2011). The function of metaplastic Paneth cells in the colon is unknown. However, these cells appear in the colon during inflammatory disease and produce a number of antimicrobial peptides such as DEFA6, DEFA5 and REG3A. Thus, the Paneth cells with their antimicrobial peptides probably have a disease-modifying action in colonic inflammation (Cunliffe et al. 2001; Shen et al. 2005). Interestingly, Cunliffe et al. (2001) have shown a similar expression pattern for DEFA5 as is seen for DEFA6, REG1B and REG3A in the present work. Of these peptides, all but REG1B have previously been demonstrated to have an antibacterial effect. Given the similarity in the sequences of REG1B and REG3A and given that their expression patterns are comparable to known antibacterial peptides such as DEFA5/6, we consider that REG1B is a good candidate for the screening of possible antimicrobial effects.
Good cell-for-cell overlap occurs between DEFA6 and REG1B/REG3A expression, with REG1B/REG3A being expressed only in the crypt base and in scattered epithelial cells of the lower crypts. REG1A shows a similar expression pattern in un-inflamed mucosa but its expression expands to almost all cells in the lower half of the colonic crypts during inflammation. This result is similar to that of an earlier study of REG1A expression by Shinozaki et al. (2001). The REG1A expression pattern suggests that newly proliferated epithelial cells have REG1A expression capacity but that they lose this capability during differentiation. This could be an effect of paracrine stimulation, whereby a signal conveyed by basal crypt cells weakens towards the lumen. This paracrine stimulation could originate in Paneth cells, based on the observation that DEFA6-positive cells are always seen in the base of crypts in which REG1A expansion to epithelial cells occurs. A Paneth-cell-dependent epithelial expression of REG could explain the high expression levels in un-inflamed samples harbouring metaplastic Paneth cells. The reported expression levels of REG1A/REG1B/REG3A in healthy small intestine could also be explained by such a mode of induction, as Paneth cells are present in healthy small bowel (Parikh et al. 2012). However, we have as yet been unable to demonstrate such a correlation unequivocally.
REG4 mRNA shows a localisation that is completely different from that of the other REGs, with expression only in scattered cells in normal mucosa, expanding to colonocytes of the upper colonic crypts during inflammation and in some un-inflamed IBD samples. The area of positive staining after expansion to colonocytes suggests that these gain REG4 expression late in differentiation. The scattered cells have been identified as enteroendocrine serotonin-producing cells. However, not all serotonin-positive cells are REG4-positive. Almost no CgA-positive/REG4-positive/serotonin-negative cells have been seen, suggesting that only the serotonin-positive subgroup of enteroendocrine cells expresses REG4 in un-inflamed mucosa. However, as several serotoninpositive/REG4-negative cells have been observed, the full nature of REG4 expression in enteroendocrine cells has not been elucidated. As reported earlier, IHC shows REG4-positive staining of the mucus excreted from goblet cells during inflammation (Granlund et al. 2011). This clearly indicates a secretory function for this protein. Earlier studies have suggested both a mitogenic and migratory effect of REG4 on colon cancer cell lines (Rafa et al. 2010). The latter observation could correlate with the expression pattern seen in the present study, with an expression gradient increasing towards the lumen, possibly stimulating colonocytes migration towards the luminal face after proliferation in the crypt base. However, little evidence supports such an effect of REG4 in non-neoplastic disease and hence further research is needed to identify the role of REG4 in the setting up of epithelial inflammation.
In conclusion, the expression pattern of REG family genes can be divided into at least two groups. REG1A, REG1B and REG3 show an apparently Paneth-cell-dependent expansion from the colonic crypts during inflammation, whereas REG4 is expressed in serotonin-producing enteroendocrine cells and expands to epithelial cells of the upper colonic crypts during inflammation. A similar expression pattern is also seen in non-IBD colonic inflammation, as represented by PC. The expression pattern of REG1A and REG1B, seen together with sequence similarities with REG3A, suggests that these peptides are good candidates for the screening of antimicrobial effects.
References
Arai N, Mitomi H, Ohtani Y, Igarashi M, Kakita A, Okayasu I (1999) Enhanced epithelial cell turnover associated with p53 accumulation and high p21WAF1/CIP1 expression in ulcerative colitis. Mod Pathol 12:604–611
Astrosini C, Roeefzaad C, Dai Y-Y, Dieckgraefe BK, Jöns T, Kemmner W (2008) REG1A expression is a prognostic marker in colorectal cancer and associated with peritoneal carcinomatosis. Int J Cancer 123:409–413
Bevins CL, Salzman NH (2011) Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9:356–368
Cash HL, Whitham CV, Behrendt CL, Hooper LV (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130
Cunliffe RN, Rose FRAJ, Keyte J, Abberley L, Chan WC, Mahida YR (2001) Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut 48:176–185
Dieckgraefe BK, Crimmins DL, Landt V, Houchen C, Anant S, Porche-Sorbet R, Ladenson JH (2002) Expression of the regenerating gene family in inflammatory bowel disease mucosa: Reg I[alpha] upregulation, processing, and antiapoptotic activity. J Invest Med 50:421–434
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang JH (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:16
Granlund AVB, Beisvag V, Torp SH, Flatberg A, Kleveland PM, Østvik AE, Waldum HL, Sandvik AK (2011) Activation of REG family proteins in colitis. Scand J Gastroenterol 46:1316–1323
Guo Y, Xu J, Li N, Gao F, Huang P (2010) RegIV potentiates colorectal carcinoma cell migration and invasion via its CRD domain. Cancer Genet Cytogenet 199:38–44
Haapamäki MM, Grönroos JM, Nurmi H, Alanen K, Kallajoki M, Nevalainen TJ (1997) Gene expression of group II phospholipase A2 in intestine in ulcerative colitis. Gut 40:95–101
Heiskala K, Giles-Komar J, Heiskala M, Andersson L (2006) High expression of RELP (Reg IV) in neoplastic goblet cells of appendiceal mucinous cystadenoma and pseudomyxoma peritonei. Virchows Arch 448:295–300
Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, Davies S, Ogunbiyi O, Nuñez G, Keshav S (2003) Crohn’s disease and the NOD2 gene: a role for Paneth cells. Gastroenterology 125:47–57
Lawrance IC, Fiocchi C, Chakravarti S (2001) Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet 10:445–456
Loftus EV Jr (2004) Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 126:1504–1517
Lomax AE, Linden DR, Mawe GM, Sharkey KA (2006) Effects of gastrointestinal inflammation on enteroendocrine cells and enteric neural reflex circuits. Auton Neurosci 126–127:250–257
Oue N, Mitani Y, Aung PP, Sakakura C, Takeshima Y, Kaneko M, Noguchi T, Nakayama H, Yasui W (2005) Expression and localization of Reg IV in human neoplastic and non-neoplastic tissues: Reg IV expression is associated with intestinal and neuroendocrine differentiation in gastric adenocarcinoma. J Pathol 207:185–198
Oue N, Kuniyasu H, Noguchi T, Sentani K, Ito M, Tanaka S, Setoyama T, Sakakura C, Natsugoe S, Yasui W (2007) Serum concentration of Reg IV in patients with colorectal cancer: overexpression and high serum levels of Reg IV are associated with liver metastasis. Oncology 72:371–380
Parikh A, Stephan AF, Tzanakakis ES (2012) Regenerating proteins and their expression, regulation, and signaling. Biomol Concepts 3:57–70
Player AN, Shen LP, Kenny D, Antao VP, Kolberg JA (2001) Single copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem 49:603–611
Rafa L, Dessein AF, Devisme L, Buob D, Truant S, Porchet N, Huet G, Buisine MP, Lesuffleur T (2010) REG4 acts as a mitogenic, motility and pro invasive factor for colon cancer cells. Int J Oncol 36:689–698
Sekikawa A, Fukui H, Suzuki K, Karibe T, Fujii S, Ichikawa K, Tomita S, Imura J, Shiratori K, Chiba T, Fujimori T (2010) Involvement of the IL 22/REG I[alpha] axis in ulcerative colitis. Lab Invest 90:496–505
Shen B, Porter EM, Reynoso E, Shen C, Ghosh D, Connor JT, Drazba J, Rho HK, Gramlich TL, Li R, Ormsby AH, Sy M-S, Ganz T, Bevins CL (2005) Human defensin 5 expression in intestinal metaplasia of the upper gastrointestinal tract. J Clin Pathol 58:687–694
Shinozaki S, Nakamura T, Iimura M, Kato Y, Iizuka B, Kobayashi M, Hayashi N (2001) Upregulation of Reg 1α and GW112 in the epithelium of inflamed colonic mucosa. Gut 48:623–629
Smyth G (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (eds) Bioinformatics and computational biology solutions using R and Bioconductor. Springer, Berlin, pp 397–420
Zelensky AN, Gready JE (2005) The C-type lectin-like domain superfamily. FEBS J 272:6179–6217
Zhang YW, Ding LS, Lai MD (2003) Reg gene family and human diseases. World J Gastroenterol 9:2635–2641
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig S1
Serial sections hybridised by using positive (Ubiquitin C, a) and negative (bacterial dapB, b) control probes. Positive controls showed strong staining, whereas negative controls demonstrated almost no unspecific binding of the probe. Bar 200 μm (JPEG 25 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License, which permits any use, distribution and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
van Beelen Granlund, A., Østvik, A.E., Brenna, Ø. et al. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res 352, 639–646 (2013). https://doi.org/10.1007/s00441-013-1592-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-013-1592-z