Abstract
The ability to extract mitochondrial and nuclear DNA from ancient remains has enabled the study of ancient DNA, a legitimate field for over 20 years now. Recently, Y chromosome genotyping has begun to be applied to ancient DNA. The Y chromosome haplogroup in East Asia has since caught the attention of molecular anthropologists, as it is one of the most ethnic-related genetic markers of the region. In this paper, the Y chromosome haplogroup of DNA from ancient East Asians was examined, in order to genetically link them to modern populations. Fifty-six human remains were sampled from five archaeological sites, primarily along the Yangtze River. Strict criteria were followed to eliminate potential contamination. Five SNPs from the Y chromosome were successfully amplified from most of the samples, with at least 62.5% of the samples belonging to the O haplogroup, similar to the frequency for modern East Asian populations. A high frequency of O1 was found in Liangzhu Culture sites around the mouth of the Yangtze River, linking this culture to modern Austronesian and Daic populations. A rare haplogroup, O3d, was found at the Daxi site in the middle reaches of the Yangtze River, indicating that the Daxi people might be the ancestors of modern Hmong-Mien populations, which show only small traces of O3d today. Noticeable genetic segregation was observed among the prehistoric cultures, demonstrating the genetic foundation of the multiple origins of the Chinese Civilization.
Similar content being viewed by others
Introduction
Ancient DNA has been studied for over 20 years, since the genetic examination of the Quagga (Higuchi et al. 1984) and an ancient Egyptian mummy (Pääbo 1985). Though the authenticity of the results from ancient DNA has always been questioned, its study has been accepted as legitimate. Strict procedures were instituted to minimize the potential for extraneous DNA contamination, and careful checks were performed to verify the authenticity of putatively ancient DNA (Pääbo 1989; Stoneking 1995; Pääbo et al. 2004). A large amount of work on ancient DNA studies has been reported, most of which were performed with mitochondrial DNA (mtDNA), since mtDNA has many more copies within each sample than nuclear DNA, increasing the opportunity for extraction. Even data from Neanderthal mtDNA, which is tens of thousands of years old, has been ascertained (Serre et al. 2004; Dalton 2006). Ancient nuclear DNA has also been studied (Lawler et al. 1991; Béraud-Colomb et al. 1995), illustrating that it is possible to obtain information from all types of DNA from ancient remains, in order to answer questions of anthropological interest.
The genetic relationship among populations of different archaeological cultures, as well as between ancient and modern populations, is of great interest to archaeologists in East Asia (Su 1999). The highly ethnic-related Y chromosome diversity is one of the best materials to describe the relationships (Su et al. 1999; Shi et al. 2005). The Y chromosome haplogroup patterns are quite different among different ethnic groups. For instance, O1 is primarily in Austronesian and Daic populations (Zhang et al. 2007). Y chromosome studies on ancient DNA were reported as well (Hummel and Herrmann 1991; Schultes et al. 1999), most of which concentrated on short tandem repeats (STRs). Two Y STR studies on East Asian ancient remains have been reported, which revealed some information of the ancient Hun’s social structure (Keyser-Tracqui et al. 2003), and the relationship between Amerindians and ancient Siberians (Ricaut et al. 2005). However, haplogroups can not be determined by Y STR data alone, but by single nucleotide polymorphisms (SNPs). The first reported ancient Y SNP data was typed from a Native American sample of an extinct tribe (Kuch et al. 2007). Another paper reported the Y SNP data of 11 ancient South Siberian samples (Bouakaze et al. 2007). In this paper, we present Y SNP data of 48 ancient samples from China, mostly along Yangtze River, to provide a survey on the genetic diversity of the prehistory populations in this region.
There were several different archaeological culture (C.) regions in East Asia in the Neolithic Age from around 9,000–3,000 years before present (Su 1999). The cultural differences among the regions lasted throughout the whole Neolithic Age and into the Bronze Age. There were two series of Neolithic cultures quite distinct from each other in the drainage area of Yangtze River. In the Three Gorges region in the middle reaches of the Yangtze River, the culture catena (CSACH 1998) consisted of Nanmuyuan C. (6000–5000 bc), Liulinxi C. (5000–4400 bc), Daxi C. (4400–3300 bc), Qujialing C. (3300–2500 bc), and Shijiahe C. Miaoping Type (2500–2200 bc). Around the mouth of the Yangtze River, the catena (ZIA 1999) consisted of Kuahuqiao C. (6000 bc∼), Majiabang C. (5100–3900 bc), Songze C. (3900–3300 bc), Liangzhu C. (3300–2100 bc), and Maqiao C. (1900–1200 bc). While the cultures of the different periods showed continuity in each catena, no evidence of intercultural communications has yet been found. In the region between the two previous regions, there was another desultory culture catena (Peng 2005): Xianrendong C. (8000 bc∼), Shinianshan C. (4000–2500 bc), Shanbei C. (2800–2000 bc), and Wucheng C. (1500–1100 bc). For comparison, there were also some developed culture catenae in the drainage area of the Yellow River in the North China. The most important was the catena of Peiligang C. (7000–5000 bc), Yangshao C. (5000–3000 bc), Longshan C. (3000–2200 bc), Erlitou C. (2200–1500 bc), and Shang C. (1500–1100 bc), which was believed to be the major origin of the Chinese Civilization. Some argue that these Neolithic cultures along the Yangtze River were also origins of Chinese Civilization. It will certainly excite the anthropologists of East Asia to reveal whether the people of the different Neolithic cultures were ancestors of modern Chinese or other nations by genetic methods. It will also raise the professional enthusiasm to know whether the culture diversification coincided with the genetic diversification.
Materials and methods
Archaeological sites and samples
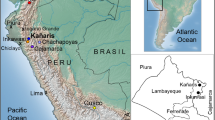
The samples collected in this study were from five sites: Maqiao, Xindili, Wucheng, Daxi, and Taosi. The locations were marked in Fig. 1. Most of the samples belonged to four different cultures: Daxi C. was the earliest, followed by the Longshan C. and Liangzhu C. around the same period, and finally, by Wucheng C., the latest in the Shang Dynasty of the Bronze Age. These cultures were most representative for the prehistory of China. Some samples from the same necropolises that belonged to the historical time (later than 841 bc in China) were also collected for comparison. Most of them belonged to the Han Dynasty. The remains of the same culture were sampled from different necropolises, avoiding the bias of relative samples.
The skeletons were buried directly in the loessal soil, with no apparent chests or coffins. Most of the skeletons excavated in the area of Yangtze River were rotten, but some of the necropolises were built in the higher places and remained in good condition, covered tightly and protected by the loess. We chose the unbroken and hardest skeletons for our samples. A loess covering of 2 in. was not removed from each skeleton before it was carried into the dedicated ancient DNA laboratory to minimize the risk of contamination during excavation and transportation. DNA of each sample was extracted and typed in one month after excavation, ensuring the freshness of the sample, which was best for amplification (Pruvost et al. 2007). The sex of each skeleton was established according to the methodology developed by Murail et al. (1999). Only male samples were subjected to the Y SNP genotyping. In total, 56 individual remains were included in the experiments. Some pieces of wood were also collected in the same way from the same tombs for extraction controls. Although animal remains would make better controls, no animal remains were found associated to the human skeleton.
Measures taken to avoid contamination and ensure authenticity
We have two isolated laboratories dedicated to work with ancient DNA. As we have mentioned, samples were never touched by any person before they were moved into the isolated labs, because they were covered by the loess in the tombs. Even when they were covered, they were handled with gloves by a reduced number of anthropologists wearing face masks and caps. During the transportation, the samples were packaged up in the plastic bags hermetically.
Our ancient DNA labs are strictly controlled following all the criteria for ancient DNA studies (Pääbo et al. 2004), such as routine sterilization by different treatments (DNAse away, positive air pressure, bleach and ultraviolet light irradiation), air filtration, and isolated rooms for different experimental steps (three rooms for sample cleaning, DNA extraction, and PCR cocktail preparation, respectively). Less than three researchers can work in each lab, wearing full body protective clothing, and using dedicated equipments and reagents. Pre-PCR work was performed in the designated rooms of the labs. PCR cocktails were prepared and sealed carefully before they were carried out of the pre-PCR rooms. The PCR room is far from the other rooms, and airflow between the PCR room and the other rooms are strictly avoided. The post-PCR productions were therefore physically separated from the pre-PCR procedures.
Only female researchers were involved in the pre-PCR procedures, preventing possible contamination from modern Y chromosome DNA. Each skeleton was sampled from several different parts: mostly teeth, astragalus, calcaneus, vertebrae, etc. The teeth we chose were all intact (without carious lesions), and still fixed to the jaw. We only used bones with thick cortical, as the density of cortical bone offers two advantages compared to spongy bone. First, the quantity of the mineral crystal of hydroxyapatite on which the DNA is fixed is higher than in the spongy bone. Second, it helps protect against contamination. The outer surface of the bones was removed to almost 2 mm of depth. The same procedures were performed in two different labs on different samples from the same skeletons for reproduction controls. For each SNP, at least three rounds of amplifications were repeated in each lab. Extraction controls were also provided. The same procedure was performed for the wood controls as for the human remains. Extraction and amplification blanks were still used as negative controls.
We did not clone the PCR products or quantify the number of starting templates in the reaction following the criteria of some researchers (Cooper and Poinar 2000; Gilbert et al. 2005), as the damage or jumping PCR will not result in mistakes in the determination of Y SNP alleles.
DNA extraction and amplification
DNA was carefully extracted according to a published protocol (Fily et al. 1998); no adaptation was made on the protocol. The sample bones were crushed under liquid nitrogen in a freezer mill and DNA was extracted by the silicone method (Gilbert et al. 2003). As the protocol has been described by many papers in brief or detail, we need not repeat it here. Extraction was performed in a clean bench hood with positive air pressure and UV irradiation, which was cleaned and sterilized between the extractions of every two samples. Only one sample was extracted at a time to avoid the cross-contamination.
The SNPs that we typed were M119, M95, M122, M7, M134. They formed five haplogroups (O1, O2a, O3*, O3d, O3e) according to the YCC nomenclature (YCC 2002). The amplification protocol was the same as that previously published (Su et al. 1999; Ke et al. 2001). The annealing circles of the PCR reactions were added to 60. The length of the PCR productions were all around 100–200 bp, shorter than the general size of ancient DNA between 100 and 500 bp (Hofreiter et al. 2001).
Results and discussion
No amplifiable product could be obtained from any of the extraction control samples (wood pieces), or from eight individual skeletons. DNA of these eight individuals might have been severely degraded (shown as missing data in Table 1). There were no differences between the non-working samples and the working samples. It was not possible for us to determine if there were wrong morphological sex determinations for the non-working samples, as the sex determination fragment of amelogenin could not be amplified for them either. For some of the samples, not all of the five SNPs could be amplified, thus haplogroups could not be determined if no mutated alleles were found (shown as undetermined in Table 1). However, most individuals were successfully amplified, and the haplogroups were determined for half of them. Therefore, at least 62.5% of the individual remains (30 out of 48) belong to O haplogroup, which is still the major haplogroup of today’s East Asians. These ancient results, consequently, did not differ from the modern populations. The resulting DNA types thus made “phylogenetic sense” (the Y chromosome haplogroup structure), helping to verify the authenticity of the ancient DNA. The previously reported results of ancient Y chromosome were all obtained from samples preserved in cold environments (Keyser-Tracqui et al. 2003; Ricaut et al. 2005; Kuch et al. 2007; Bouakaze et al. 2007). However, our samples were not buried in a really cold environment. Therefore, the relatively high amplification rate should have resulted from the strict selection of the best-preserved remains.
In two sites of Liangzhu Culture, only O1 haplogroup were found, and the frequencies of O1 in two sites were almost the same. That indicated the ancient people of these two sites belonged to the same population. The historical samples from the same sites did not differ from the prehistoric samples in the haplogroup patterns. Even modern populations in this area around Shanghai contain a large proportion of O1 haplogroup (Wen et al. 2004). The consistency of the Y haplogroup pattern in this area, from the Neolithic Age to modern times, reveals that the population might not have been replaced. O1 reaches the highest frequencies in the Taiwan aborigines, and also in Daic speaking populations in the southwest China (Li 2005). Therefore, it is also possible that there are some close relationships among Taiwan aborigines, Daic speakers and the ancient Liangzhu Culture populations.
A high frequency of O3d was only found in Daxi Culture. O3d is very rare in modern populations; Hmong-Mien populations have been found to contain a small proportion of O3d (Feng 2007). Among those Hmong-Mien populations, She and Bunu were found to have the highest frequency of O3d (Su et al. 1999). Since O3d occurs at low frequency in the Hmong-Mien, the ancient people of Daxi Culture might be the ancestors of the modern populations of Hmong-Mien. The absence of O3d in the historical samples from the Daxi site (it might not have been found because of the small sample size), and the migration of modern Hmong-Mien populations to the southwest might indicate that the prehistoric population in the Three Gorges area has been replaced.
Among three sites—Wucheng, Daxi and Taosi—O3* and O2a were shared. As O3* is most common in modern East Asians, and O2a can been found in different populations in southwestern China, we can not resolve the close relationships or the genetic flows among the populations of these sites, according to the shared haplogroups. There were still some genetic differences between the populations in the areas of Yangtze River and the Yellow River, as O2a was not found from the Taosi site.
O1 was not found out of Liangzhu Culture, bringing a noticeable genetic difference between coastal and inland populations, which can still be observed in modern populations. O1 distributes along the coast of East Asia, from Manchuria in the north to Malaysia and Indonesia in the south (Su et al. 1999; Li 2005; Zhang et al. 2007). This distribution may indicate that there were at least two different migration routes for the early peopling of East Asia. Hardly any genetic flows could be observed between the coastal route and the inland route in the prehistoric peoples.
In conclusion, the genetic diversities among the prehistoric people in several regions of East Asia indicated that different archaeological culture catenae were founded by the people of different origins. Genetic segregations might occur much earlier than the diversification of the Neolithic cultures, and were not broken until those prehistoric cultures mixed to form the Chinese Civilization.
References
Béraud-Colomb E, Roubin R, Martin J, Maroc N, Gardeisen A, Trabuchet G, Goossens M (1995) Human P-globin gene polymorphisms characterized in DNA extracted from ancient bones 12,000 years old. Am J Hum Genet 57:1267–1274
Bouakaze C, Keyser C, Amory S, Crubézy E, Ludes B (2007) First successful assay of Y-SNP typing by SNaPshot minisequencing on ancient DNA. Int J Legal Med (in press)
Chinese State Administration of Culture Heritage (1998) Discovery of archaeology in Three Gorges: the third excavation at Wushan Daxi Site. Hubei Science and Technology Press, Wuhan
Cooper A, Poinar HN (2000) Ancient DNA: do it right or not at all. Science 289:1139
Dalton R (2006) Neanderthal DNA yields to genome foray. Nature 441(7091):260–261
Feng H (2007) General history of Yao Nationality (Yaozu Tongshi). Minzu Press, Beijing
Fily ML, Crubézy E, Courtaud P, Keyser C, Ebrard D, Ludes B (1998) Analyse paléogénétique des sujets de la grotte sépulcrale d’Elzarreko Karbia (Bronze ancien, Pays Basque). CR Acad Sci III(321):79–85
Gilbert MT, Willerslev E, Hansen AJ, Barnes I, Rudbeck L, Lynnerup N, Cooper A (2003) Distribution patterns of postmortem damage in human mitochondrial DNA. Am J Hum Genet 72:32–47
Gilbert MT, Bandelt HJ, Hofreiter M, Barnes I (2005) Assessing ancient DNA studies. Trends Ecol Evol 20:541–544
Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC (1984) DNA sequences from the quagga, an extinct member of the horse family. Nature 312:282–284
Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S (2001) Ancient DNA. Nat Rev Genet 2:353–359
Hummel S, Herrmann B (1991) Y-chromosome-specific DNA amplified in ancient human bone. Naturwissenschaften 78(6):266–267
Ke Y, Su B, Xiao J, Chen H, Huang W, Chen Z, Chu J, Tan J, Jin L, Lu D (2001) Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians. Sci China C Life Sci 44:225–232
Keyser-Tracqui C, Crubezy E, Ludes B (2003) Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia. Am J Hum Genet 73(2):247–260
Kuch M, Grocke DR, Knyf MC, Gilbert MT, Younghusband B, Young T, Marshall I, Willerslev E, Stoneking M, Poinar H (2007) A preliminary analysis of the DNA and diet of the extinct Beothuk: a systematic approach to ancient human DNA. Am J Phys Anthropol 132(4):594–604
Lawlor DA, Dickel CD, Hauswirth WW, Parham P (1991) Ancient HLA genes from 7,500-year-old archaeological remains. Nature 349(6312):785–788
Li H (2005) Genetic structure of Austro-Tai populations. Ph.D. dissertation of Human Biology, Fudan University, Shanghai
Murail P, Bruzek J, Braga J (1999) A new approach to sexual diagnosis in past populations: practical adjustments from Van Vark’s procedure. Int J Osteoarchaeol 9:39–53
Pääbo S (1985) Molecular cloning of ancient Egyptian mummy DNA. Nature 314:644–645
Pääbo S (1989) Ancient DNA; extraction, characterization, molecular cloning and enzymatic amplification. Proc Natl Acad Sci USA 86:1939–1943
Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M (2004) Genetic analyses from ancient DNA. Annu Rev Genet 38:645–679
Peng M-h (2005) Culture research about Wucheng. Wenwu Press, Beijing
Pruvost M, Schwarz R, Correia VB, Champlot S, Braguier S, Morel N, Fernandez-Jalvo Y, Grange T, Geigl EM (2007) Freshly excavated fossil bones are best for amplification of ancient DNA. Proc Natl Acad Sci USA 104(3):739–744
Ricaut FX, Fedoseeva A, Keyser-Tracqui C, Crubezy E, Ludes B (2005) Ancient DNA analysis of human neolithic remains found in northeastern Siberia. Am J Phys Anthropol 126(4):458–462
Schultes T, Hummel S, Herrmann B (1999) Amplification of Y-chromosomal STRs from ancient skeletal material. Hum Genet 104:164–166
Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S (2004) No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol 2(3):E57
Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B (2005) Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet 77(3):408–419
Stoneking M (1995) Ancient DNA: how do you know when you have it and what can you do with it? Am J Hum Genet 57(6):1259–1262
Su BQ (1999) New survey on the origin of Chinese Civilization. SDX Joint Publishing Co, Beijing
Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724
The Y Chromosome Consortium (2002) A nomenclature system of the tree of human Y-chromosomal binary haplogroup. Genome Res 12:339–348
Wen B, Li H, Lu DR, Song XF, Zhang F, He YG, Li F, Gao Y, Mao XY, Zhang L, Qian J, Tan JZ, Jin JZ, Huang W, Deka R, Su B, Chakraborty R, Jin L (2004) Genetic evidence supports demic diffusion of Han culture. Nature 431:302–305
Zhang F, Su B, Zhang YP, Jin L (2007) Genetic studies of human diversity in East Asia. Philos Trans R Soc Lond B Biol Sci 362:987–995
Zhejiang Institute of Archaeology (1999) Liangzhu Culture researches. Science Press, Beijing
Acknowledgments
The authors thank the CPAM of Shanghai, Zhejiang Institute of Archaeology, Jiangxi Institute of Archaeology and Shanxi Institute of Archaeology for providing the samples.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, H., Huang, ., Mustavich, L.F. et al. Y chromosomes of prehistoric people along the Yangtze River. Hum Genet 122, 383–388 (2007). https://doi.org/10.1007/s00439-007-0407-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-007-0407-2