Abstract
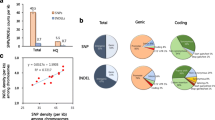
Genetic diversity within parental lines of hybrid rice is the foundation of heterosis utilization and yield improvement. Previous studies have suggested that genetic diversity was narrow in cytoplasmic male sterile (CMS/A line) and restorer lines (R line) for Three-line hybrid rice. However, the genetic diversity within maintainer lines (B line), especially at a genome-wide scale, remains largely unknown. In the present study, we performed deep re-sequencing of the elite maintainer line V20B (Oryza sativa L. ssp. indica). We then compared the V20B sequence with the 93-11 (Oryza sativa L. ssp. indica) genome sequence. 112.1 × 106 paired-end reads (PE reads) were generated with approximately 30-fold sequencing depth. The V20B PE reads uniquely covered 87.6 % of the 93-11 genome sequence. Overall, a total of 660,778 single-nucleotide polymorphism (SNPs) and 266,301 insertions and deletions (InDels) were identified, yielding an average of 2.1 SNPs/kb and 0.8 InDels/kb. Genome-wide distribution of the SNPs and InDels was non-random, and variation-rich and variation-poor regions were identified in all chromosomes. A total of 20,562 non-synonymous SNPs spanning 8,854 genes were annotated. Our results identified DNA polymorphisms at the genome-wide scale and uncovered the high level of genetic diversity between V20B and 93-11. Our results proved that next-generation sequencing technologies can be powerful tools to study genome-wide DNA polymorphisms, to query genetic diversity, and to enable molecular improvement efforts with Three-line hybrid rice. Further, our results also indicated that 93-11 could be used as core germplasm for the improvement of wild-abortive CMS lines and the maintainer lines.






Similar content being viewed by others
Abbreviations
- SNPs:
-
Single-nucleotide polymorphisms
- InDels:
-
Short insertions and deletions
- SVs:
-
Structural variations
- CMS:
-
Cytoplasmic male sterile
- nsSNPs:
-
Non-synonymous SNPs
- NGS:
-
Next-generation sequencing
- MAS:
-
Marker-assisted selection
- QTL:
-
Quantitative trait loci
References
Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, Wakasa K (2011) Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol 52:274–282
Batley J, Barker G, O’Sullivan H, Edwards KJ, Edwards D (2003) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 132:84–91
Bentley DR (2006) Whole-genome re-sequencing. Curr Opin Genet Dev 16:545–552
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314
Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL et al (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet 3:1745–1756
Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15:57–71
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Ersoz ES, Yu J, Buckler ES (2007) Applications of linkage disequilibrium and association mapping in crop plants. Genomics-assisted crop improvement. Springer, The Netherlands, pp 97–119
Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14:1812–1819
Ganal MW, Altmann T, Röder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12:211–217
Gao Q, Yue G, Li W, Wang J, Xu J, Yin Y (2012) Recent progress using high-throughput sequencing technologies in plant molecular breeding. J Integr Plant Biol 54:215–227
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp.japonica). Science 296:92–100
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A et al (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17:388–390
Jones N, Ougham H, Thomas H, Pašakinskienė I (2009) Markers and mapping revisited: finding your gene. New Phytol 183:935–966
Li R, Fan W, Tian G, Zhu H, He L, Cai J et al (2009a) The sequence and de novo assembly of the giant panda genome. Nature 463:311–317
Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J (2009b) SNP detection for massively parallel whole-genome re-sequencing. Genome Res 19:1124–1132
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009c) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li S, Wang S, Deng Q, Zheng A, Zhu J, Liu H et al (2012) Identification of genome-wide variations among three elite restorer lines for hybrid-rice. PLoS One 7:e30952
Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, Namiki N et al (2005) The map-based sequence of the rice genome. Nature 436:793–800
McCouch SR, Zhao K, Wright M, Tung CW, Ebana K, Thomson M et al (2010) Development of genome-wide SNP assays for rice. Breed Sci 60:524–535
Morton BR (1995) Neighboring base composition and transversion/transition bias in a comparison of rice and maize chloroplast noncoding regions. Proc Natl Acad Sci USA 92:9717–9721
Nagasaki H, Ebana K, Shibaya T, Yonemaru JI, Yano M (2010) Core single-nucleotide polymorphisms-a tool for genetic analysis of the Japanese rice population. Breed Sci 60:648–655
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW (1998) Microarrays: biotechnology’s discovery platform for functional genomics. Trends Biotechnol 16:301–306
Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY et al (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135:1198–1205
Subbaiyan GK, Waters DL, Katiyar SK, Sadananda AR, Vaddadi S, Henry RJ (2012) Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol J 10:623–634
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98:9161–9166
Varshney RK, Hoisington DA, Tyagi AK (2006) Advances in cereal genomics and applications in crop breeding. Trends Biotechnol 24:490–499
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27:522–530
Wang L, Hao L, Li X, Hu S, Ge S, Yu J (2009) SNP deserts of Asian cultivated rice: genomic regions under domestication. J Evol Biol 22:751–761
Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X et al (2012) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30:105–111
Yamamoto T, Nagasaki H, Yonemaru JI, Ebana K, Nakajima M, Shibaya T, Yano M (2010) Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics 11:267
Yu J, Hu S, Wang J, Wong GKS, Li S, Liu B et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Yu H, Xie W, Wang J, Xing Y, Xu C, Li X et al (2011) Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS One 6:e17595
Yuan LP, Virmani SS (1988) Status of hybrid rice research and development. International Rice Research Institute, Los Baños, pp 7–24 (Hybrid rice. Manila)
Yuan LP, Yang ZY, Yang JB (1994) Hybrid rice research in China. In Hybrid rice technology: New developments and future prospects. Selected papers from the International Rice Research Conference, Manila, pp 143–148
Acknowledgments
We thank the staffs of the Beijing Genomics Institute, Shenzhen, for helping with data analysis, and we thank Zhicheng Yuan and Wuzhun Luo of the China National Hybrid Rice Research and Development Center for management of our field plantings.
This research was financially supported by the earmarked fund for Modern Agro-industry Technology Research system, Genetically modified organisms breeding major projects (2013ZX08001-004) and Science and technology Department of Hunan province science and technology plan key projects (2012FJ2005).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The experiments comply with the current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Bigang Mao contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1. General statistics of predicted protein-coding genes of V20B unmapped reads.
Supplementary Table S2. Statistics of function annotation.
Supplementary Table S3. Validation of SNPs by Sanger method.
Supplementary Table S4. Validation of insertions and deletions by Sanger method.
Supplementary Table S5. Regions with high- or low-density of SNPs.
Supplementary Table S6. Regions with high- or low-density of InDels.
Supplementary Table S7. Annotation of SNPs in gene region.
Supplementary Table S8. The length distribution and ratio of insertions and deletions.
Supplementary Table S9. Annotation of InDels in gene region.
Supplementary Table S10. Functional annotation of the 809 genes.
Supplementary material. Sets of SNPs and InDels.
Rights and permissions
About this article
Cite this article
Hu, Y., Mao, B., Peng, Y. et al. Deep re-sequencing of a widely used maintainer line of hybrid rice for discovery of DNA polymorphisms and evaluation of genetic diversity. Mol Genet Genomics 289, 303–315 (2014). https://doi.org/10.1007/s00438-013-0807-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-013-0807-z