Abstract
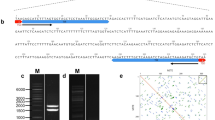
Short INterspersed Elements (SINEs) in invertebrates, and especially in animal inbred genomes such that of termites, are poorly known; in this paper we characterize three new SINE families (Talub, Taluc and Talud) through the analyses of 341 sequences, either isolated from the Reticulitermes lucifugus genome or drawn from EST Genbank collection. We further add new data to the only isopteran element known so far, Talua. These SINEs are tRNA-derived elements, with an average length ranging from 258 to 372 bp. The tails are made up by poly(A) or microsatellite motifs. Their copy number varies from 7.9 × 103 to 105 copies, well within the range observed for other metazoan genomes. Species distribution, age and target site duplication analysis indicate Talud as the oldest, possibly inactive SINE originated before the onset of Isoptera (~150 Myr ago). Taluc underwent to substantial sequence changes throughout the evolution of termites and data suggest it was silenced and then re-activated in the R. lucifugus lineage. Moreover, Taluc shares a conserved sequence block with other unrelated SINEs, as observed for some vertebrate and cephalopod elements. The study of genomic environment showed that insertions are mainly surrounded by microsatellites and other SINEs, indicating a biased accumulation within non-coding regions. The evolutionary dynamics of Talu~ elements is explained through selective mechanisms acting in an inbred genome; in this respect, the study of termites’ SINEs activity may provide an interesting framework to address the (co)evolution of mobile elements and the host genome.





Similar content being viewed by others
References
Akasaki T, Nikaido M, Nishihara H, Tsuchiya K, Segawa S, Okada N (2010) Characterization of a novel SINE superfamily from invertebrates: “Ceph-SINEs” from the genomes of squids and cuttlefish. Gene 454:8–19
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Calleri DV II, McGrail Reid E, Rosengaus RB, Vargo EL, Traniello JFA (2006) Inbreeding and disease resistance in a social insect: effects of heterozygosity on immunocompetence in the termite Zootermopsis angusticollis. Proc R Soc B 273:2633–2640
Diaz-Gonzalez J, Dominguez A, Albornoz J (2010) Genomic distribution of retrotransposons 297, 1731, copia, mdg1 and roo in the Drosophila melanogaster species subgroup. Genetica 138:579–586
Dolgin ES, Charlesworth B (2006) The fate of transposable elements in asexual populations. Genetics 174:817–827
Engel MS, Grimaldi DA, Krishna K (2009) Termites (Isoptera): their phylogeny, classification, and rise to ecological dominance. Am Mus Novitat 3650:1–27
Feschotte C, Fourrier N, Desmons I, Mouchès C (2001) Birth of a retroposon: the Twin SINE family from the vector mosquito Culex pipiens may have originated from a dimeric tRNA precursor. Mol Biol Evol 18:74–84
Gilbert N, Labuda D (1999) CORE-SINEs: eukaryotic short interspersed retroposing elements with common sequence motifs. PNAS 96:2869–2874
Gilbert N, Labuda D (2000) Evolutionary inventions and continuity of CORE-SINEs in mammals. J Mol Biol 298:365–377
Gogolevsky KP, Vassetzky NS, Kramerov DA (2009) 5S rRNA-derived and tRNA-derived SINEs in fruit bats. Genomics 93:494–500
Goodier JL, Kazazian HH Jr (2008) Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23–31
Holt RA, Subramanian GM, Halpern A et al (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science 298:129–149
Hua-Van A, Le Rouzic A, Maisonhaute C, Capy P (2005) Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet Genome Res 110:426–440
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Jurka J, Kapitonov VV, Kohany O, Jurka MV (2007) Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genom Human Genet 8:241–259
Kajikawa M, Okada N (2002) LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111:433–444
Koshikawa S, Miyazaki S, Cornette R, Matsumoto T, Miura T (2008) Genome size of termites (Insecta, Dictyoptera, Isoptera) and wood roaches (Insecta, Dictyoptera, Cryptocercidae). Naturwissenschaften 95:859–867
Kramerov DA, Vassetzky NS (2005) Short retroposons in eukaryotic genomes. Int Rev Cytol 247:165–221
Le Rouzic A, Boutin TS, Capy P (2007) Long-term evolution of transposable elements. PNAS 104:19375–19380
Lenoir A, Lavie L, Prieto J-L, Goubely C, Côté J-C, Pélissier T, Deragon J-M (2001) The evolutionary origin and genomic organization of SINEs in Arabidopsis thaliana. Mol Biol Evol 18:2315–2322
Li Y-C, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Luchetti A (2005) Identification of a short interspersed repeat in Reticulitermes lucifugus (Isoptera Rhinotermitidae) genome. DNA Seq 16:304–307
Luchetti A, Mantovani B (2009) Talua SINE biology in the genome of the Reticulitermes subterranean termites (Isoptera, Rhinotermitidae). J Mol Evol 69:589–600
Luchetti A, Marini M, Mantovani B (2006) Non-concerted evolution of the RET76 satellite DNA family in Reticulitermes taxa (Insecta, Isoptera). Genetica 128:123–132
Mayer C (2009) Phobos v. 3.3.11, 2006–2010. Ruhr-Universität Bochum. http://www.rub.de/spezzoo/cm/cm_phobos.htm. Accessed 12 July 2010
Mita K, Kasahara M, Sasaki S et al (2004) The genome sequence of silkworm, Bombyx mori. DNA Res 11:27–35
Nishihara H, Okada N (2008) Retroposons: genetic footprints on the evolutionary paths of life. In: Murphy WJ (ed) Methods in molecular biology: phylogenomics. Humana Press Inc, Totowa, pp 201–225
Nishihara H, Smit FAA, Okada N (2006) Functional noncording sequences derived from SINEs in the mammalian genome. Genome Res 16:864–874
Ogiwara I, Miya M, Oshima K, Okada N (2002) V-SINEs: a new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Res 12:316–324
Ohshima K, Okada N (2005) SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res 110:475–490
Piskurek O, Okada N (2007) Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. PNAS 104:12046–12051
Silva JC, Loreto EL, Clark JB (2004) Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol 6:57–72
Sunter JD, Patel SP, Skilton RA, Githaka N, Knowles DP, Scoles GA, Nene V, de Villiers E, Bishop RP (2008) A novel SINE family occurs frequently in both genomic DNA and transcribed sequences in ixodid ticks of the arthropod sub-phylum Chelicerata. Gene 415:13–22
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
The International Aphid Genomics Consortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8:e1000313
Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955
Tu Z (1999) Genomic and evolutionary analysis of Feilai, a diverse family of highly reiterated SINEs in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol 16:760–766
Tu Z (2001) Maque, a family of extremely short interspersed repetitive elements: characterization, possible mechanism of transposition, and evolutionary implication. Gene 263:247–253
Tu Z, Li S, Mao C (2004) The changing tails of a novel short interspersed element in Aedes aegypti: genomic evidence for slippage retrotransposition and the relationship between 3′ tandem repeats and the poly(dA) tail. Genetics 168:2037–2047
Vargo EL, Husseneder C (2009) Biology of subterranean termites: insights from molecular studies of Reticulitermes and Coptotermes. Annu Rev Entomol 54:379–403
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:873–982
Wright SI, Schoen DJ (1999) Transposon dynamics and the breeding system. Genetica 107:139–148
Acknowledgments
This work has been supported by Canziani donation, Fondazione del Monte—Bologna and PRIN-2008 funds to BM. We are particularly grateful to Claudia Husseneder for her encouragement to use the Coptotermes formosanus EST library she produced. We also wish to thank the two anonymous referees whose suggestions substantially improved this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luchetti, A., Mantovani, B. Molecular characterization, genomic distribution and evolutionary dynamics of Short INterspersed Elements in the termite genome. Mol Genet Genomics 285, 175–184 (2011). https://doi.org/10.1007/s00438-010-0595-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-010-0595-7