Abstract
The nonautonomous nDart1 element in the hAT superfamily is one of a few active DNA transposons in rice. Its transposition can be induced by crossing with a line containing an active autonomous element, aDart1, and stabilized by segregating aDart1. No somaclonal variation should occur in nDart1-promoted gene tagging because no tissue culture is involved in nDart1 activation. By transposon display analysis, we examined the activities of nDart1-related elements in the selfed progeny of a mutable virescent pyl-v plant containing aDart1. Although various nDart1-related elements are present in the rice genome, only nDart1-3 subgroup elements, nDart1-0 and nDart1-3 in particular, were found to be transposed frequently and integrated into various sites almost all over the genome, and a fraction of the transposed elements were found to be transmitted to the next generation. More than half of the newly integrated elements were identified as nDart1-0. Analysis of the newly inserted sites revealed that the nDart1-3 subgroup elements were predominantly integrated into single-copy regions. More than 60% of the transposed elements were inserted into the genic regions that comprise putative coding regions and their 0.5-kb flanking segments, and approximately two-thirds of them were within the 0.5-kb area in front of the putative initiation codons, i.e., promoter-proximal genic regions. These characteristic features of nDart1-3 subgroup elements seem to be suitable for developing an efficient and somaclonal variation-free gene tagging system for rice functional genomics.







Similar content being viewed by others
References
An S, Park S, Jeong D-H, Lee D-Y, Kang H-G, Yu J-H, Hur J, Kim S-R, Kim Y-H, Lee M, Han S, Kim S-J, Yang J, Kim E, Wi SJ, Chung HS, Hong J-P, Choe V, Lee H-K, Choi J-H, Nam J, Kim S-R, Park P-B, Park KY, Kim WT, Choe S, Lee C-B, An G (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133:2040–2047
Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36:105–113
Chopra S, Hoshino A, Boddu J, Iida S (2006) Flavonoid pigments as tools in molecular genetics. In: Glotewold E (ed) The science of flavonoids. Springer, New York, pp 147–173
Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Genet Genomics 273:150–157
Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4:118–122
Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, Sundaresan V, Leung H (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54:325–334
Huang J, Zhang K, Shen Y, Huang Z, Li M, Tang D, Gu M, Cheng Z (2009) Identification of a high frequency transposon induced by tissue culture, nDaiZ, a member of the hAT family in rice. Genomics 93:274–281
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jeong D-H, An S, Park S, Kang H-G, Park G-G, Kim S-R, Sim J, Kim Y-O, Kim M-K, Kim S-R, Kim J, Shin M, Jung M, An G (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45:123–132
Johzuka-Hisatomi Y, Maekawa M, Takagi K, Eun C-H, Yamauchi T, Shimatani Z, Ahmed N, Urawa H, Tsugane K, Terada R, Iida S (2008) Homologous recombination-dependent gene targeting and an active DNA transposon nDart-promoted gene tagging for rice functional genomics. In: Hirano H-Y, Hirai A, Sano Y, Sasaki T (eds) Rice biology in the genomics era: biotechnology in agriculture and forestry, vol 62. Springer, Berlin, pp 81–94
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188
Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun B, Park SH, Park JY, Lee EJ, Kim MJ, Chung WS, Lee KH, Lee YS, Lee JJ, Won YJ, Yi G, Nam MH, Cha YS, Yun DW, Eun MY, Han C-D (2004) Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J 39:252–263
Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37:301–314
Kumar CS, Wing RA, Sundaresan V (2005) Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J 44:879–892
Kunze R, Saedler H, Lönnig W-E (1997) Plant transposable elements. Adv Bot Res 27:332–470
Leung H, An G (2004) Rice functional genomics: large-scale gene discovery and applications to crop improvement. Adv Agron 82:55–111
Maekawa M, Rikiishi K, Matsuura T, Noda K (1999) A marker line H-126, carries a genetic factor making chlorophyll mutation variegated. Rice Genet Newslett 16:61–62
Maekawa M, Tsugane K, Iida S (2010) Effective contribution of the nDart transposon-tagging system to rice functional genomics. Adv Genet Res 4 (in press)
May BP, Martienssen RA (2003) Transposon mutagenesis in the study of plant development. Crit Rev Plant Sci 22:1–35
Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15:1771–1780
Miyao A, Iwasaki Y, Kitano H, Itoh J-I, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H (2007) A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol 63:625–635
Moon S, Jung K-H, Lee D-E, Jiang W-Z, Koh HJ, Heu M-H, Lee DS, Suh HS, An G (2006) Identification of active transposon dTok, a member of the hAT family, in rice. Plant Cell Physiol 47:1473–1483
Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103:17620–17625
Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–1134
Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T (2003) Mobilization of a transposon in the rice genome. Nature 421:170–172
Nishimura H, Ahmed N, Tsugane K, Iida S, Maekawa M (2008) Distribution and mapping of an active autonomous aDart element responsible for mobilizing nonautonomous nDart1 transposons in cultivated rice varieties. Theor Appl Genet 116:395–405
Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, Piegu B, Droc G, Regad F, Bourgeois E, Meynard D, Perin C, Sabau X, Ghesquiere A, Glaszmann JC, Delseny M, Guiderdoni E (2004) High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J 39:450–464
Satoh K, Doi K, Nagata T, Kishimoto N, Suzuki K et al (2007) Gene organization in rice revealed by full-length cDNA mapping and gene expression analysis through microarray. PLoS ONE 2:e1235
Shimatani Z, Takagi K, Eun C-H, Maekawa M, Takahara H, Hoshino A, Qian Q, Terada R, Johzuka-Hisatomi Y, Iida S, Tsugane K (2009) Characterization of autonomous Dart1 transposons belonging to the hAT superfamily in rice. Mol Genet Genomics 281:329–344
Takagi K, Ishikawa N, Maekawa M, Tsugane K, Iida S (2007) Transposon display for active DNA transposons in rice. Genes Genet Syst 82:109–122
The Rice Annotation Project (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17:175–183
The Rice Full-Length cDNA Consortium (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301:376–379
Tsugane K, Maekawa M, Takagi K, Takahara H, Qian Q, Eun C-H, Iida S (2006) An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J 45:46–57
Upadhyaya NM (2007) Rice functional genomics—challenges, progress and prospects. Springer, New York
van Enckevort LJG, Droc G, Piffanelli P, Greco R, Gagneur C, Weber C, Gonzalez VM, Cabot P, Fornara F, Berri S, Miro B, Lan P, Rafel M, Capell T, Puigdomenech P, Ouwerkerk PBF, Meijer AH, Pe E, Colombo L, Christou P, Guiderdoni E, Pereira A (2005) EU-OSTID: a collection of transposon insertional mutants for functional genomics in rice. Plant Mol Biol 59:99–110
Acknowledgments
We thank Yoshio Sano for his encouragement, Miwako Matsumoto, Yoko Kobayashi, Seiko Nakano, and Kazue Hiramatsu for their technical assistance, Atsushi Hoshino for reading the manuscript, and Chang-Ho Eun for helpful discussions. This work was supported by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) from Bio-oriented Technology Research Advancement Institution (BRAIN) in Japan (to S.I. and M.M.) and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 17207002 to S.I. and 22780007 to K.T.). It was also partly supported by the Global COE Program (to S.I.) and the NIBB Cooperative Research Program (9-153 to M.M). K. Takagi is a recipient of a fellowship awarded by the Japan Society for the Promotion of Science for Young Scientists (No. 04J09255).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by M.-A. Grandbastien.
Electronic supplementary material
Below is the link to the electronic supplementary material.
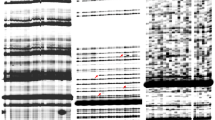
Supplementary Fig. 1.
nDart-TD and nDart1-0-TD analyses in Nipponbare and several R plants. Lanes with NP and numerals indicate DNA samples from Nipponbare and from the samples used in Fig. 5, respectively, and the same nDart-TD and nDart1-0-TD procedures as in Fig. 5 are employed. Nine nDart1-3 subgroup elements detected in both Nipponbare and pyl-v are indicated by filled arrowheads, whereas the two elements, nDart1-0 and nDart1-3(3-3), found only in pyl-v are indicated by open arrowheads. The symbols nDart1-0 [pyl-v], nDart1-0 [Chr 2], and nDart1-0 [Chr 6] indicate nDart1-0 residing at the pyl-v locus and at the long arms of chromosomes 2 and 6, respectively (PDF 527 kb)
Rights and permissions
About this article
Cite this article
Takagi, K., Maekawa, M., Tsugane, K. et al. Transposition and target preferences of an active nonautonomous DNA transposon nDart1 and its relatives belonging to the hAT superfamily in rice. Mol Genet Genomics 284, 343–355 (2010). https://doi.org/10.1007/s00438-010-0569-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-010-0569-9