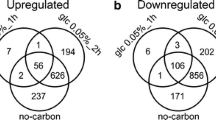
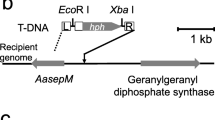
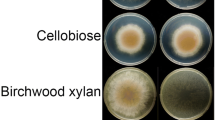
Abstract
Hexokinases catalyse the first step in glucose metabolism and play a role in glucose sensing in mammals, plants and fungi. We describe a new class of hexokinases that appear to be solely regulatory in function. The Aspergillus nidulans hxkD gene (formerly named xprF) encodes a hexokinase-like protein. We constructed hxkDΔ gene disruption mutants which showed increased levels of extracellular protease in response to carbon starvation. The hxkDΔ mutations are not completely recessive, indicating that the level of the gene product is critical. Transcript levels of hxkD increase during carbon starvation and this response is not dependent on functional HxkD. A gene encoding a second atypical hexokinase (HxkC) was identified. The hxkCΔ gene disruption mutant exhibits a phenotype similar, but not identical, to hxkDΔ mutants. As with hxkD, mutations in hxkC are suppressed by loss-of-function mutations in xprG, which encodes a putative transcriptional activator involved in the response to nutrient limitation. We show that GFP-tagged HxkD was found only in nuclei suggesting a regulatory role for HxkD. GFP-tagged HxkC was associated with mitochondria. Homologs of hxkC and hxkD are conserved in multi-cellular fungi. Genes encoding atypical hexokinases are present in many genome sequence databases. Thus, non-catalytic hexokinases may be widespread.






Similar content being viewed by others
References
Bali D, Svetlanov A, Lee H-W, Fusco-DeMane D, Leiser M, Li B, Barzilai N, Surana M, Hou H, Fleischer N, DePinho R, Rossetti L, Efrat S (1995) Animal model for maturity-onset diabetes of the young generated by disruption of the mouse glucokinase gene. J Biol Chem 270:21464–21467
Bork P, Sander C, Valencia (1993) Convergent evolution of similar enzymatic function on different protein folds: the hexokinase ribokinase, and galactokinase families of sugar kinases. Protein Sci 2:31–40
Clutterbuck AJ (1974) Aspergillus nidulans. In: King RC (ed), Handbook of genetics, vol 1. Plenum Press, New York, pp 447–510
Clutterbuck AJ (1993) Aspergillus nidulans (nuclear genes). In: O’Brien SJ (ed) Locus maps of complex genomes: lower eukaryotes, vol 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 3.71–3.84
Cohen BL (1973) Regulation of intracellular and extracellular neutral and alkaline proteases in Aspergillus nidulans. J Gen Microbiol 79:311–320
Copeley SD (2003) Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol 7:265–272
Cove DJ (1966) The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta 113:51–56
Cubero B, Scazzocchio C (1994) Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J 13:407–415
Daniel J (2005) Sir-dependent downregulation of various aging processes. Mol Genet Genomics 274:539–547
Davies RW (1994) Heterologous gene expression and protein secretion in Aspergillus. In: Martinelli SD, Kinghorn JR (eds) Aspergillus: 50 years on. Progress in industrial microbiology, vol 29. Elsevier Science BV, Amsterdam, pp 527–560
Dementhon K, Iyer G, Glass NL (2006) VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora. Eukaryot Cell, published online ahead of print
Efrat S, Tal M, Lodish HF (1994) The pancreatic beta-cell glucose sensor. Trends Biochem Sci 19:535–538
Entian K-D, Fröhlich K-U (1984) Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol 158:29–35
Enyenihi AH, Saunders WS (2003) Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47–54
Felsenstein J (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. In: Doolittle RF (ed) Methods in enzymology, vol 266. Academic, San Diego, pp 418–427
Fidel S, Doonan JH, Morris NR (1988) Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283–293
Flipphi M, van de Vondervoort PJI, Ruijter GJG, Visser J, Arst HN Jr, Felenbok B (2003) Onset of carbon catabolite repression in Aspergillus nidulans: parallel involvement of hexokinase and glucokinase in sugar signalling. J Biol Chem 278:11849–11857
Fröhlich K-U, Entian K-D, Mecke D (1985) The primary structure of the yeast hexokinase PII gene (HXK2) which is responsible for glucose repression. Gene 36:105–111
Hohmann S, Winderickx J, de Winde JH, Valckx D, Cobbaert P, Luyten K, de Meirsman C, Ramos J, Thevelein JM (1999) Novel alleles of yeast hexokinase PII with distinct effects on catalytic activity and catabolite repression SUC2. Microbiol 145:703–714
Katz ME, Hynes MJ (1989) Characterization of the amdR-controlled lamA and lamB genes of Aspergillus nidulans. Genetics 122:331–339
Katz ME, Flynn PK, vanKuyk P, Cheetham BF (1996) Mutations affecting extracellular protease production in the filamentous fungus, Aspergillus nidulans. Mol Gen Genet 250:715–724
Katz ME, Masoumi A, Burrows SR, Shirtliff CG, Cheetham BF (2000) The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of the extracellular proteases. Genetics 156:1559–1571
Katz ME, Gray K–A, Cheetham BF (2006) The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet Biol 43:190–199
Kiemer AD, Takeuchi K, Quinlan MP (2001) Identification of genes involved in epithelial-mesenchymal transition and tumor progression. Oncogene 20:6679–6688
Kim M, Lim J-H, Ahn CS, Park K, Kim GT, Kim WT, Pai H-S (2006) Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana bethamiana. Plant Cell 18:2341–2355
Kochetov AV, Ischenko IV, Vorobriev DG, Kel AE, Babenko VN, Kisselev LL, Kolchanov NA (1998) Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett 440:351–355
Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B (1993) Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol 7:847–857
Kuser PR, Krauchenco S, Antunes OAC, Polikarpov I (2000) The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J Biol Chem 275:20814–20821
Maitra PK, Lobo Z (1983) Genetics of yeast glucokinase. Genetics 105:501–515
Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K, Chandel NS, Thompson CB, Robey RB, Hay N (2004) Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell 16:819–830
Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300:332–336
Nicholas KB (2001) GeneDoc 2.6.02 [Online] http://www.psc.edu/biomed/genedoc
Oakley CE, Weil CF, Kretz PL, Oakley BR (1987) Cloning of the riboB locus of Aspergillus nidulans. Gene 53:293–298
Panneman H, Ruijter GJG, van den Broeck HC, Driever ETM, Visser J (1996) Cloning and biochemical characterization of an Aspergillus niger glucokinase. Evidence for the presence of separate glucokinase and hexokinase enzymes. Eur J Biochem 240:518–525
Panneman H, Ruijter GJG, van den Broeck HC, Visser J (1998) Cloning and biochemical characterization of Aspergillus niger hexokinase. The enzyme is strongly inhibited by physiological concentrations of trehalose 6-phosphate. Eur J Biochem 258:223–232
Panozzo C, Cornillot E, Felenbok B (1998) The CreA repressor is the sole DNA-binding protein responsible for carbon catabolite repression of the alcR gene in Aspergillus nidulans via its binding to a couple of specific sites. J Biol Chem 273:6367–6372
Pastorino JG, Shulga N, Hoek JB (2002) Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem 277:7610–7618
Pego JV, Weisbeek PJ, Smeekens SCM (1999) Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol 119:1017–1023
Punt PJ, Dingemanse MA, Kuyvenhoven J, Soede RDM, Pouwels PH, van den Hondel CAMJJ (1990) Functional elements in the promoter region of the Aspergillus nidulans gpdA gene coding for glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109
Ravagnani A, Gorfinkiel L, Langdon T, Diallinas G, Adjadj E, Demais S, Gorton D, Arst HN Jr, Scazzocchio C (1997). Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J 16:3974–3986
Reinert WR, Patel VB, Giles NH (1981) Genetic regulation of the qa gene cluster of Neurospora crassa: induction of qa messenger ribonucleic acid and dependency on qa-1 function. Mol Cell Biol 1:829–835
Rolland F, Winderickx J, Thevelein JM (2001) Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci 26:310–317
Ruijter GJG, Panneman H, van den Broeck HC, Bennett JM, Visser J (1996) Characterization of the Aspergillus nidulans frA1 mutant: hexose phosphorylation and apparent lack of involvement of hexokinase in glucose repression. FEMS Microbiol Lett 139:223–228
Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2:410–418
Shi Y, Shi Y (2004) Metabolic enzymes and coenzymes in transcription-a direct link between metabolism and transcription? Trends Genet 20:445–452
Small AJ, Todd RB, Zanker MC, Delimitrou S, Hynes MJ, Davis MA (2001) Functional analysis of TamA, a coactivator of nitrogen-related gene expression in Aspergillus nidulans. Mol Genet Genomics 265:636–646
Stoffel M, Frougel Ph, Takeda J, Zouali H, Vionnet N, Nishi S, Weber IT, Harrison RW, Pilkis SJ, Lesage S, Vaxillaire M, Velho G, Sun F, Iris F, Passa Ph, Cohen D, Bell GI (1992) Human glucokinase gene: Isolation, characterisation and identification of two missense mutations linked to early-onset non-insulin-dependent (type 2) diabetes mellitus. Proc Natl Acad Sci USA 89:7698–7702
Strauss J, Horvath HK, Abdallah BM, Kindermann J, Mach RL, Kubicek CP (1999) The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol 32:169–178
Szewczyk E, Andrianopoulos A, Davis MA, Hynes MJ (2001) A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADP-dependent isocitrate dehydrogenase in Aspergillus nidulans. J Biol Chem 276:37722–37729
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tilburn J, Scazzocchio C, Taylor GG, Zabicky-Zissman JH, Lockington RA, Davies RW (1983) Transformation by integration in Aspergillus nidulans. Gene 26:205–221
Unkles SE (1992) Gene organization in industrial filamentous fungi. In: Kinghorn JR, Turner G (eds) Applied molecular genetics of filamentous fungi. Blackie Academic and Professional, Chapman and Hall, Glasgow, pp 28–53
Upshall A, Gilbert T, Saari G, O’Hara PJ, Weglenski P, Perse B, Miller K, Timberlake WE (1986) Molecular analysis of the argB gene of Aspergillus nidulans. Mol Gen Genet 204:349–354
Walsh RB, Clifton D, Horak J, Fraenkel DG (1991) Saccharomyces cerevisiae null mutants in glucose phosphorylation: metabolism and invertase expression. Genetics 128:521–527
Wilson JE (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206:2049–2057
Yanagisawa S, Yoo S-D, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425:521–525
Zhao J, Hyman LL, Moore C (1999) Formation of mRNA 3′ ends in Eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA Synthesis. Microbiol Mol Biol Rev 63:405–445
Acknowledgments
This work was supported by the Australian Research Council and a University of New England Research Scholarship for S.M.H. Bernardo. We gratefully acknowledge E. Griffiths for construction of the HxkD expression plasmid, A. Andrianopoulos for providing pALX215, M. J. Hynes, M. Davis, and A. Andrianopoulos for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Fischer.
Electronic supplementary material
Below are the links to the electronic supplementary materials.
Fig7
FIG. S1 (A) The 5’ and (B) 3’-UTRs of the hxkD gene. The hxkD coding region has been presented previously (Katz et al. 2000), and the numbering of nucleotides shown here is relative to the first base of the start codon. The major transcriptional start site as determined by 5’-RACE is marked with an inverted triangle. A putative TATA sequence is in bold and underlined twice. Sequences similar to the consensus sequence for CAAT boxes (Unkles 1992) are written in bold and underscored. The intron in the 5’-UTR is written in lowercase letters and the splice-junctions and branch site are underscored. Circles are placed above sequences which, on either strand, conform to the CreA-binding sequence, SYGGRG (Kulmburg et al. 1993; Cubero and Scazzocchio 1994; Pannozzo et al. 1998). Filled-circles are placed above sequences which conform to the AreA-binding sequence, HGATAR (Ravagnani et al. 1997). The end of the transcript as determined by 3’-RACE is marked with a filled inverted-triangle. The restriction enzyme sites PstI and HindIII which mark the ends of the sequence that is carried by the plasmid pMK370 (Katz et al. 2000) are boxed.
Fig8
FIG. S2. Alignment of the HxkC and HxkD amino acid sequences of A. nidulans (An, AN4255.2 and AN2180.2, www.broad.mit.edu/annotation/fungi/aspergillus/) with homologs from N. crassa (Nc, NCU04728.1 and NCU06996.1, www.broad.mit.edu/annotation/fungi/neurospora/), Magnaporthe grisea (Mg, MG00623.4 and MG01424.4, www.broad.mit.edu/annotation/fungi/magnaporthe/) and Fusarium graminearum (Fg, FG08399.1 and FG09808.1, www.broad.mit.edu/annotation/fungi/fusarium/). The sequences of the well-characterised S. cerevisiae (Sc) hexokinase PII (Frölich et al. 1985), the A. nidulans hexokinase (HxkA) and A. nidulans glucokinase (GlkA) are included for comparison. Amino acids that are conserved in all 11 proteins are shaded in black, in at least 80% of the proteins in dark gray and in at least 40% of the proteins in light gray. The ATP-binding sites and sugar-binding sites, identified by Bork et al. (1993), are marked with lines above the sequence. Residues that are conserved or similar in hexokinases possessing catalytic activity are indicated with asterisks. The putative nuclear localization sequences in the A. nidulans HxkC and HxkD sequences are underlined. The alignment was generated with the CLUSTAL W (Thompson et al. 1994) and the GeneDoc (Nicholas 2001) computer programs.
Rights and permissions
About this article
Cite this article
Bernardo, S.M.H., Gray, KA., Todd, R.B. et al. Characterization of regulatory non-catalytic hexokinases in Aspergillus nidulans . Mol Genet Genomics 277, 519–532 (2007). https://doi.org/10.1007/s00438-006-0203-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0203-z